Crystal structure of theophylline aptamer obtained in the presence of caffeine
Kondo, J., Ohtani, Y.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| Theophylline aptamer | 33 | synthetic construct | 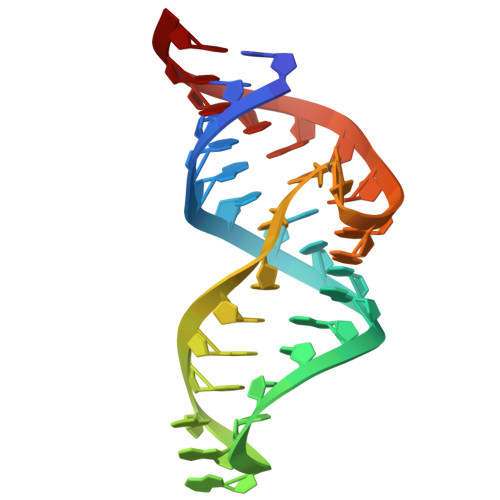 | ||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NCO Query on NCO | B [auth A] C [auth A] D [auth A] E [auth A] F [auth A] | COBALT HEXAMMINE(III) Co H18 N6 DYLMFCCYOUSRTK-UHFFFAOYSA-N |  | ||
| K Query on K | H [auth A] | POTASSIUM ION K NPYPAHLBTDXSSS-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 46.925 | α = 90 |
| b = 46.925 | β = 90 |
| c = 180.258 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XSCALE | data scaling |
| XDS | data reduction |
| AutoSol | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Japan Agency for Medical Research and Development (AMED) | Japan | JP23ama121014 |