Substrate-engaged TIM23 complex from yeast
Yang, Y.Q., Wang, G.P.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Mitochondrial import inner membrane translocase subunit TIM17 | 158 | Saccharomyces cerevisiae S288C | Mutation(s): 0 Gene Names: TIM17, MIM17, MPI2, SMS1, YJL143W, J0648 | 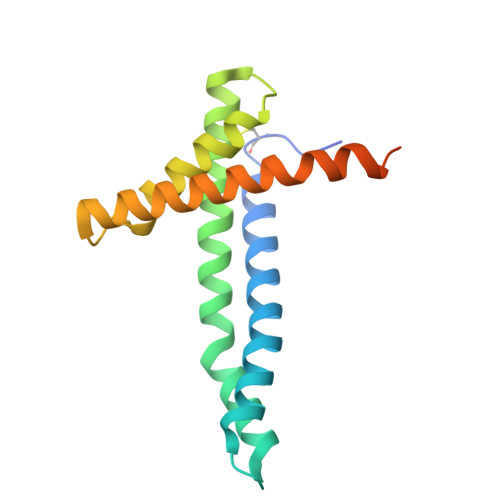 | |
UniProt | |||||
Find proteins for P39515 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P39515 Go to UniProtKB: P39515 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P39515 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Mitochondrial import inner membrane translocase subunit TIM23 | 222 | Saccharomyces cerevisiae S288C | Mutation(s): 0 Gene Names: TIM23, MAS6, MIM23, MPI3, YNR017W, N3180 | 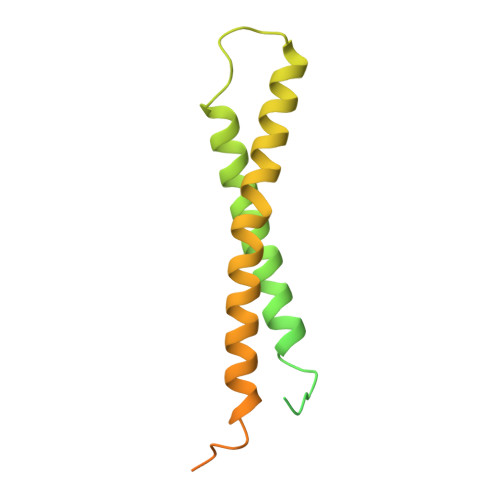 | |
UniProt | |||||
Find proteins for P32897 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P32897 Go to UniProtKB: P32897 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P32897 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Mitochondrial import inner membrane translocase subunit TIM44,L-lactate dehydrogenase (cytochrome),sfGFP | 757 | Saccharomyces cerevisiae S288C, synthetic construct | Mutation(s): 0 Gene Names: TIM44, ISP45, MIM44, MPI1, YIL022W, CYB2, YML054C, YM9958.08C EC: 1.1.2.3 | 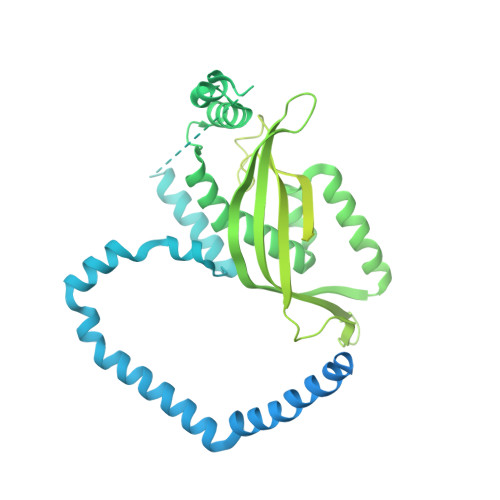 | |
UniProt | |||||
Find proteins for P00175 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P00175 Go to UniProtKB: P00175 | |||||
Find proteins for Q01852 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore Q01852 Go to UniProtKB: Q01852 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Groups | Q01852P00175 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein MGR2 | 113 | Saccharomyces cerevisiae S288C | Mutation(s): 0 Gene Names: MGR2, YPL098C | 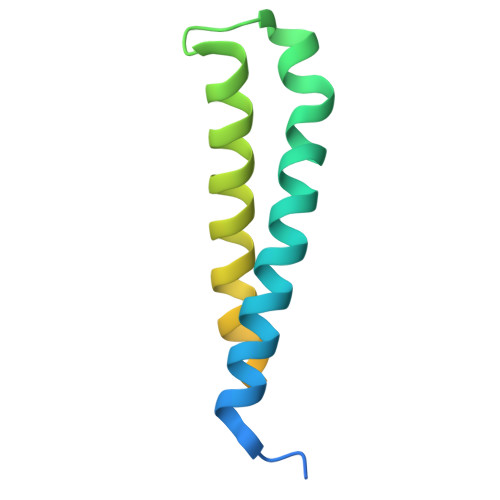 | |
UniProt | |||||
Find proteins for Q02889 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore Q02889 Go to UniProtKB: Q02889 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q02889 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| anti-Tim44-Fab H chain | E [auth H] | 238 | Saccharomyces cerevisiae S288C | Mutation(s): 0 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| anti-Tim44-Fab L chain | F [auth L] | 238 | Saccharomyces cerevisiae S288C | Mutation(s): 0 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Substrate | G [auth E] | 15 | Saccharomyces cerevisiae S288C | Mutation(s): 0 |  |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CDL (Subject of Investigation/LOI) Query on CDL | H [auth A] | CARDIOLIPIN C81 H156 O17 P2 XVTUQDWPJJBEHJ-KZCWQMDCSA-L |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| RECONSTRUCTION | cryoSPARC |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | 32271269 |