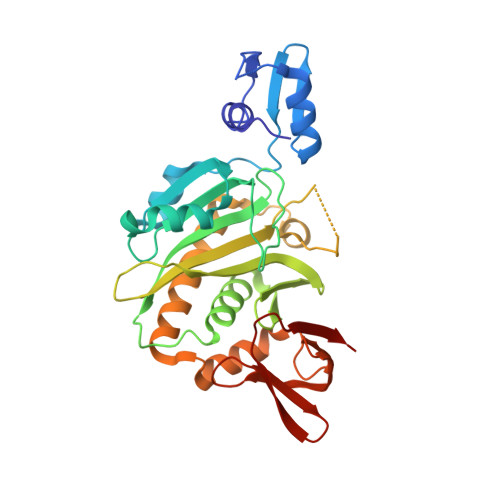
Structural insights into BirA from Haemophilus influenzae, a bifunctional protein as a biotin protein ligase and a transcriptional repressor.
Jeong, K.H., Son, S.B., Ko, J.H., Lee, M., Lee, J.Y.(2024) Biochem Biophys Res Commun 733: 150601-150601
- PubMed: 39213703
- DOI: https://doi.org/10.1016/j.bbrc.2024.150601
- Primary Citation of Related Structures:
9J8E, 9J8F - PubMed Abstract:
Biotin is an essential coenzyme involved in various metabolic processes across all known organisms, with biotinylation being crucial for the activity of carboxylases. BirA from Haemophilus influenzae is a bifunctional protein that acts as a biotin protein ligase and a transcriptional repressor. This study reveals the crystal structures of Hin BirA in both its apo- and holo-(biotinyl-5'-AMP bound) forms. As a class II BirA, it consists of three domains: N-terminal DNA binding domain, central catalytic domain, and C-terminal SH3-like domain. The structural analysis shows that the biotin-binding loop forms an ordered structure upon biotinyl-5'-AMP binding. This facilitates its interaction with the ligand and promotes protein dimerization. Comparative studies with other BirA homologs from different organisms indicate that the residues responsible for binding biotinyl-5'-AMP are highly conserved. This study also utilized AlphaFold2 to model the potential heterodimeric interaction between Hin BirA and biotin carboxyl carrier protein, thereby providing insights into the structural basis for biotinylation. These findings enhance our understanding of the structural and functional characteristics of Hin BirA, highlighting its potential as a target for novel antibiotics that disrupt the bacterial biotin synthesis pathways.
- Department of Life Science, Dongguk University-Seoul, Ilsandong-gu, Goyang-si, Gyeonggi-do, 10326, Republic of Korea.
Organizational Affiliation: