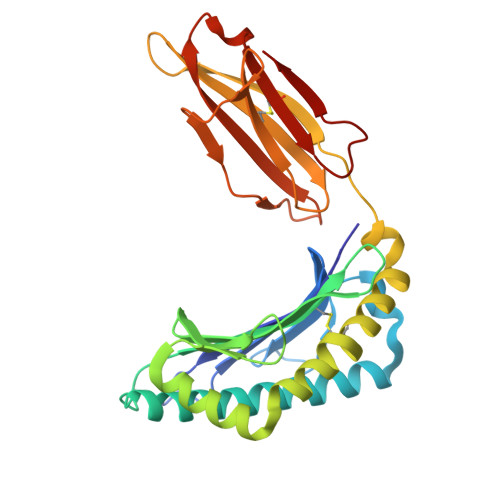
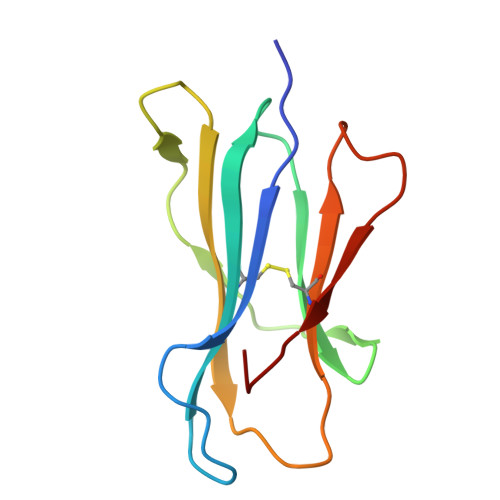
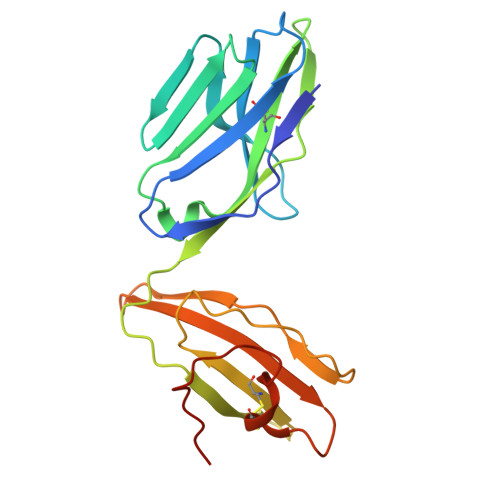
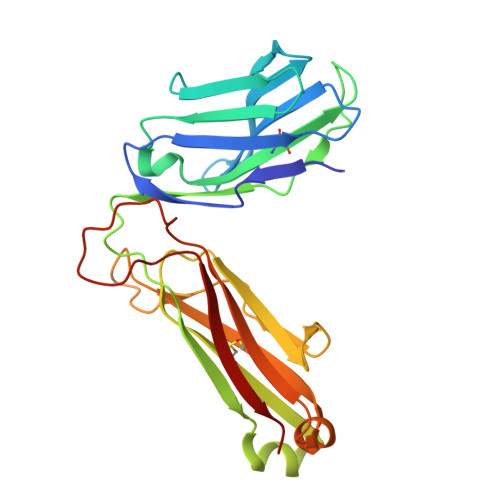
Structural insights into clonal restriction and diversity in T cell recognition of two immunodominant SARS-CoV-2 nucleocapsid epitopes.
Yuan, P., Chen, G., Li, Y., Liu, X., Saravanakumar, S., Zhao, J., Ji, Q., Wang, H., Lin, Y.W., Elbahnasawy, M., Weng, N.P., Pierce, B.G., Mariuzza, R.A., Wu, D.(2025) Nat Commun 16: 11457-11457
- PubMed: 41372155
- DOI: https://doi.org/10.1038/s41467-025-66322-6
- Primary Citation of Related Structures:
9J4T, 9J4U, 9J4V, 9WBD - PubMed Abstract:
T cells play a crucial role in clearing SARS-CoV-2 and in forming long-term memory responses to that coronavirus. The highly immunogenic nucleocapsid (N) protein of SARS-CoV-2 is much more conserved than the spike (S) protein across variants of concern, making it an attractive vaccine target for activating cytotoxic CD8 + T cells. Of particular interest are the immunodominant N epitopes LLL and SPR. Whereas LLL elicits a clonally restricted T cell response, the response to SPR is highly diverse. To understand the basis for this difference, here we determine structures of T cell receptors (TCRs) bound to LLL-HLA-A2 and SPR-HLA-B7, revealing the structural underpinnings of highly restricted Vα gene usage by LLL-specific TCRs, as well as multiple structural solutions to recognizing SPR and thereby generating a clonally diverse T cell response to that epitope. These structures also provide frameworks for understanding T cell recognition of SARS-CoV-2 variants and other coronaviruses. Finally, we compare the X-ray structures of TCR-LLL-HLA-A2 and TCR-SPR-HLA-B7 complexes with models predicted by multiple versions of AlphaFold, highlighting some success while showing room for improvement. Overall, our findings expand understanding of coronavirus T cell recognition, informing vaccine design and advances in computational modeling approaches.
- Department of Hepatopancreatobiliary Surgery, The First Affiliated Hospital, Hengyang Medical School, University of South China, Hengyang, Hunan, China.
Organizational Affiliation: