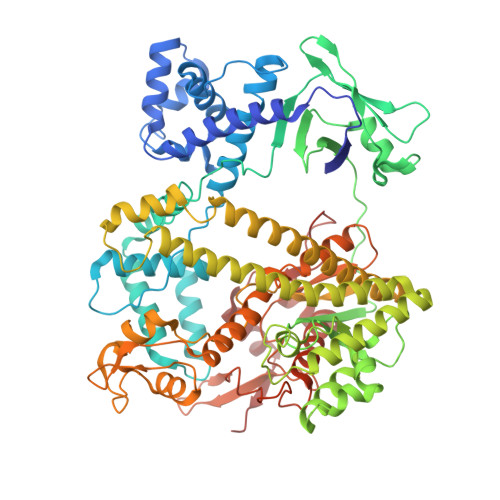
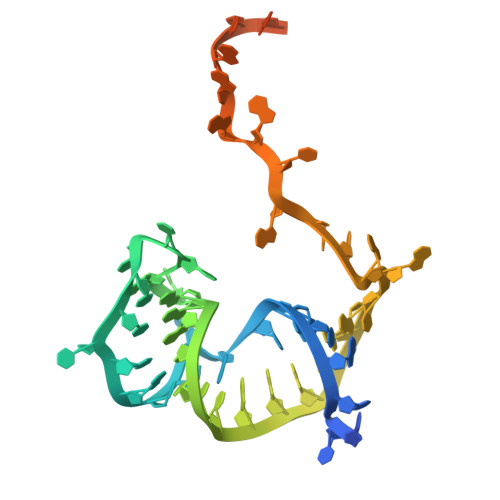
Structural basis for target DNA cleavage and guide RNA processing by CRISPR-Cas lambda 2.
Omura, S.N., Alfonse, L.E., Ornstein, A., Morinaga, H., Hirano, H., Itoh, Y., Munoz, G., Garrity, A.J., Hoffman, G.R., DiTommaso, T., Yan, W.X., Cheng, D.R., Scott, D.A., Maben, Z., Nureki, O.(2025) Commun Biol 8: 876-876
- PubMed: 40473912
- DOI: https://doi.org/10.1038/s42003-025-08300-8
- Primary Citation of Related Structures:
9IZM, 9IZP, 9IZQ, 9IZR, 9IZS - PubMed Abstract:
RNA-guided CRISPR-Cas nucleases are widely used as versatile genome-engineering tools. Among the diverse CRISPR-Cas effectors, CRISPR-Casλ-also referred to as Cas12n-is a recently identified miniature type V nuclease encoded in phage genomes. Given its demonstrated nuclease activity in both mammalian and plant cells, Casλ has emerged as a promising candidate for genome-editing applications. However, the precise molecular mechanisms of Casλ family enzymes remain poorly understood. In this study, we report the identification and detailed biochemical and structural characterizations of CRISPR-Casλ2. The cryo-electron microscopy structures of Casλ2 in five different functional states unveiled the dynamic domain rearrangements during its activation. Our biochemical analyses indicated that Casλ2 processes its precursor crRNA to a mature crRNA using the RuvC active site through a unique ruler mechanism, in which Casλ2 defines the spacer length of the mature crRNA. Furthermore, structural comparisons of Casλ2 with Casλ1 and CasΦ highlighted the diversity and conservation of phage-encoded type V CRISPR-Cas enzymes. Collectively, our findings augment the mechanistic understanding of diverse CRISPR-Cas nucleases and establish a framework for rational engineering of the CRISPR-Casλ-based genome-editing platform.
- Department of Biological Sciences, Graduate School of Science, The University of Tokyo, Tokyo, Japan.
Organizational Affiliation: