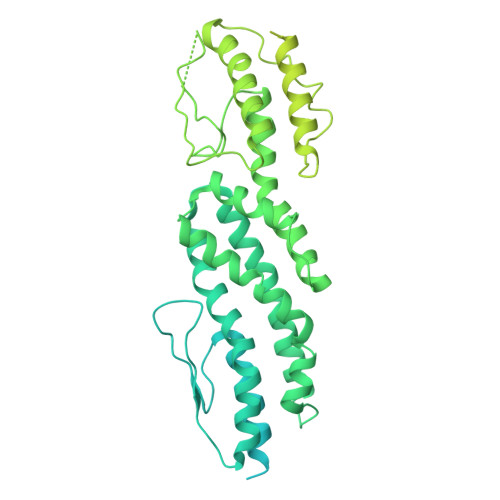
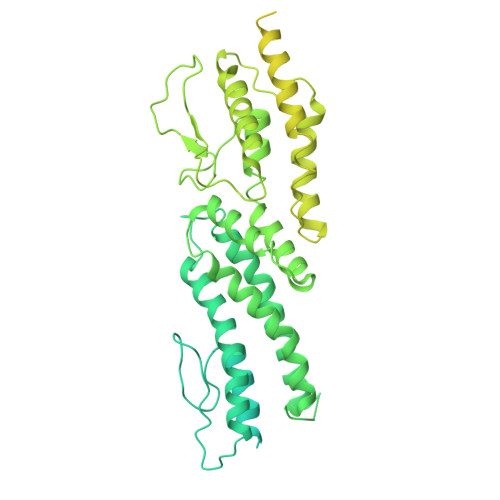
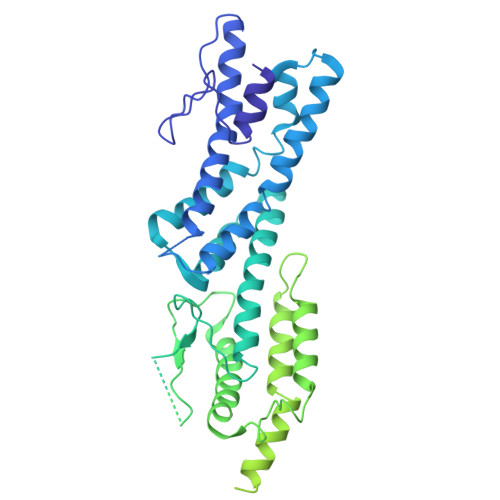
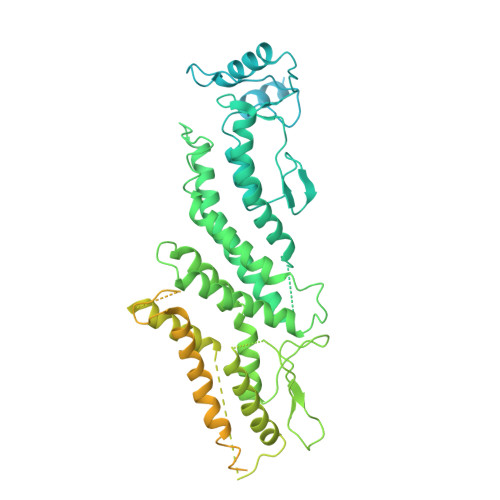
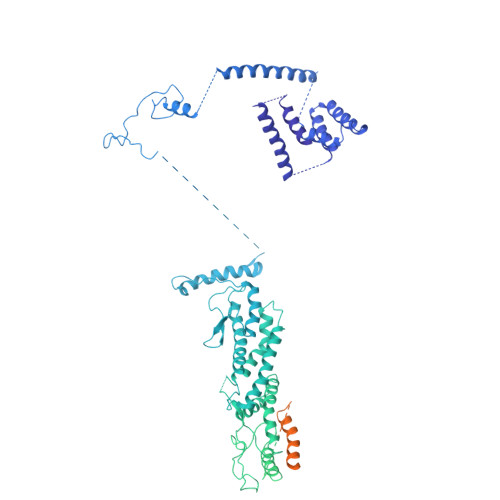
Structural mechanisms for centrosomal recruitment and organization of the microtubule nucleator gamma-TuRC.
Gao, Q., Hofer, F.W., Filbeck, S., Vermeulen, B.J.A., Wurtz, M., Neuner, A., Kaplan, C., Zezlina, M., Sala, C., Shin, H., Gruss, O.J., Schiebel, E., Pfeffer, S.(2025) Nat Commun 16: 2453-2453
- PubMed: 40074789
- DOI: https://doi.org/10.1038/s41467-025-57729-2
- Primary Citation of Related Structures:
9I8G, 9I8H, 9I8M, 9I8N - PubMed Abstract:
The γ-tubulin ring complex (γ-TuRC) acts as a structural template for microtubule formation at centrosomes, associating with two main compartments: the pericentriolar material and the centriole lumen. In the pericentriolar material, the γ-TuRC is involved in microtubule organization, while the function of the centriole lumenal pool remains unclear. The conformational landscape of the γ-TuRC, which is crucial for its activity, and its centrosomal anchoring mechanisms, which determine γ-TuRC activity and turnover, are not understood. Using cryo-electron tomography, we analyze γ-TuRCs in human cells and purified centrosomes. Pericentriolar γ-TuRCs simultaneously associate with the essential adapter NEDD1 and the microcephaly protein CDK5RAP2. NEDD1 forms a tetrameric structure at the γ-TuRC base through interactions with four GCP3/MZT1 modules and GCP5/6-specific extensions, while multiple copies of CDK5RAP2 engage the γ-TuRC in two distinct binding patterns to promote γ-TuRC closure and activation. In the centriole lumen, the microtubule branching factor Augmin tethers a condensed cluster of γ-TuRCs to the centriole wall with defined directional orientation. Centriole-lumenal γ-TuRC-Augmin is protected from degradation during interphase and released in mitosis to aid chromosome alignment. This study provides a unique view on γ-TuRC structure and molecular organization at centrosomes and identifies an important cellular function of centriole-lumenal γ-TuRCs.
- Zentrum für Molekulare Biologie der Universität Heidelberg (ZMBH), Heidelberg, Germany.
Organizational Affiliation: