Fine antigenic mapping of human enterovirus 71 from structures of cognate complexes for a panel of 12 patient derived monoclonal antibodies
Zhou, D., Ren, J., Stuart, D.I.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| light chain | A [auth L], C [auth A] | 216 | Homo sapiens | Mutation(s): 0 | 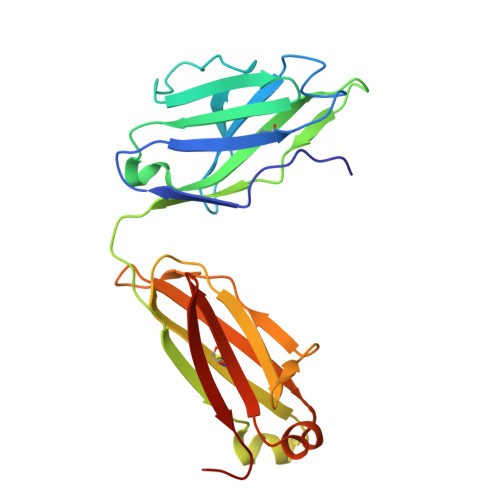 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| heavy chain | B [auth H], D [auth B] | 235 | Homo sapiens | Mutation(s): 0 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| IMD (Subject of Investigation/LOI) Query on IMD | I [auth H] | IMIDAZOLE C3 H5 N2 RAXXELZNTBOGNW-UHFFFAOYSA-O |  | ||
| EDO (Subject of Investigation/LOI) Query on EDO | E [auth L] J [auth H] K [auth H] N [auth H] P [auth B] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| FMT (Subject of Investigation/LOI) Query on FMT | F [auth L] G [auth L] H [auth L] L [auth H] M [auth H] | FORMIC ACID C H2 O2 BDAGIHXWWSANSR-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 75.319 | α = 90 |
| b = 83.945 | β = 103.5 |
| c = 84.955 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| MOLREP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Medical Research Council (MRC, United Kingdom) | United Kingdom | MR/N00065X/1 |
| Wellcome Trust | United Kingdom | 090532/Z/09/Z |