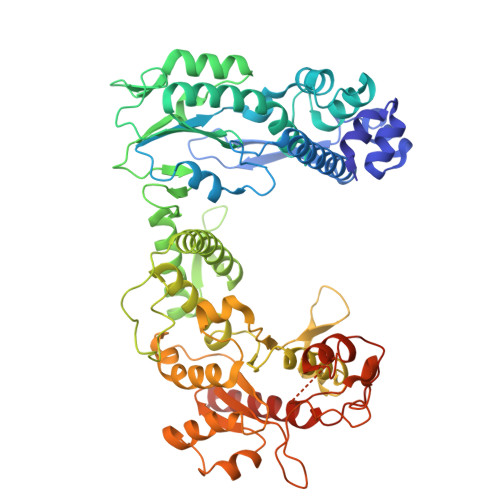
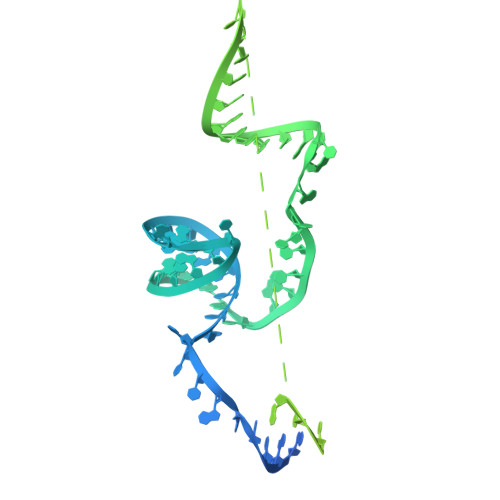
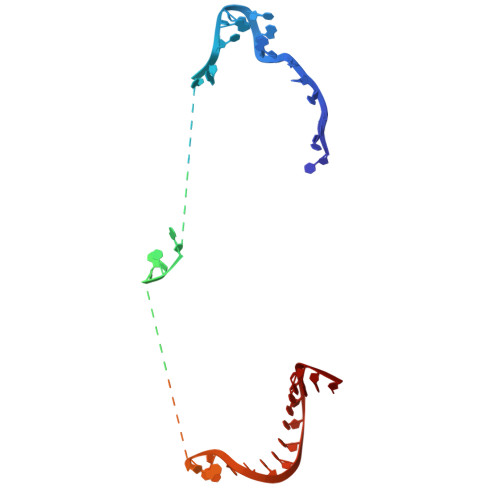
Retron Eco2 breaks tRNAs for anti-phage defense
Jasnauskaite, M., Juozapaitis, J., Liegute, T., Grigaitis, R., Skorupskaite, A., Steinchen, W., Miksys, M., Truncaite, L., Kazlauskaite, K., Khochare, S., Torres Jimenez, M.F., Dudas, G., Bange, G., Malinauskaite, L., Songailiene, I., Pausch, P.To be published.