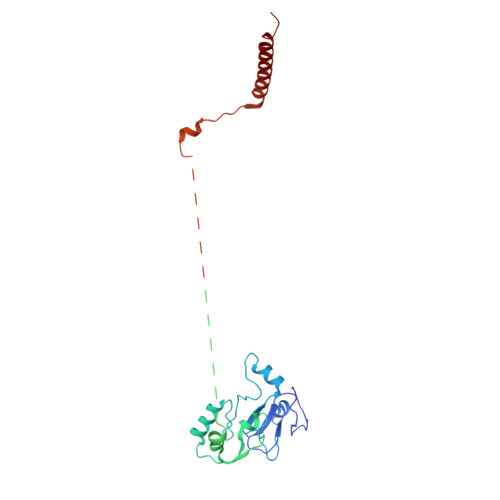
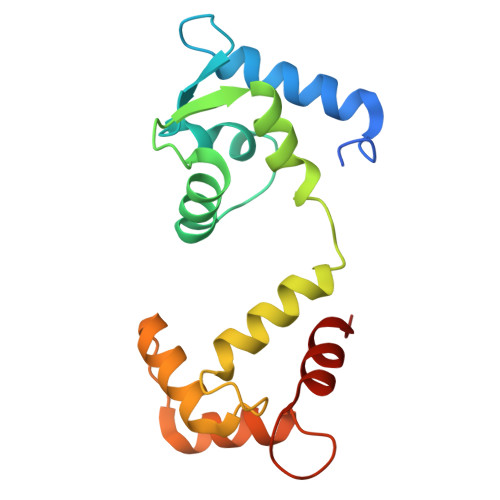
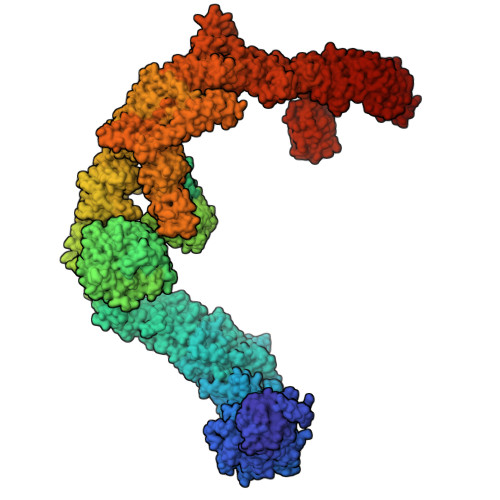
Architecture of the UBR4 complex, a giant E4 ligase central to eukaryotic protein quality control
Grabarczyk, D.B., Ehrmann, J.F., Murphy, P., Kurzbauer, R., Bell, L.E., Deszcz, L., Neuhold, J., Schleiffer, A., Shulkina, A., Versteeg, G.A., Meinhart, A., Zavodszky, E., Hegde, R.S., Clausen, T.To be published.