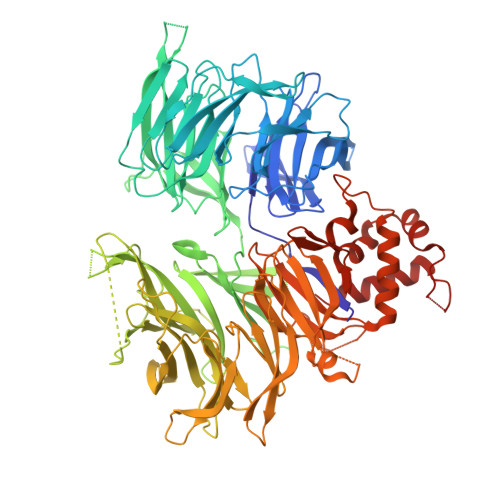
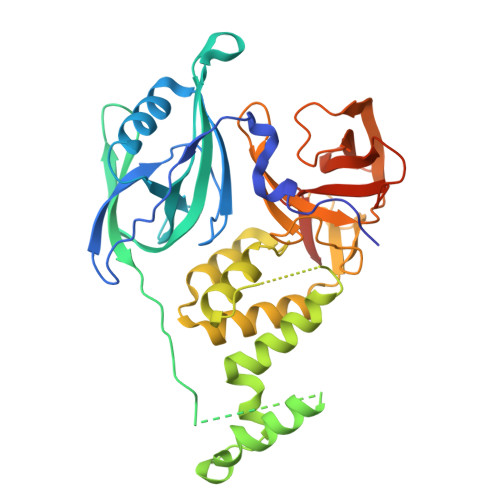
A degron-mimicking molecular glue drives CRBN homo-dimerization and degradation.
Langousis, G., Gainza, P., Hunkeler, M., Kapsitidou, D., Donckele, E.J., Annunziato, S., Wiedmer, L., Jones, K.F.M., DeMarco, B., Quan, C., Bunker, R.D., Lumb, K.J., Fasching, B., Castle, J.C., Townson, S.A., Bonenfant, D.(2025) Nat Commun 16: 10157-10157
- PubMed: 41258141
- DOI: https://doi.org/10.1038/s41467-025-65094-3
- Primary Citation of Related Structures:
9HPI, 9HPJ - PubMed Abstract:
Cereblon (CRBN) is an E3 ubiquitin ligase widely harnessed for targeted protein degradation (TPD). We report the discovery of a molecular glue degrader (MGD), MRT-31619, that drives homo-dimerization of CRBN and promotes its fast, potent, and selective degradation by the ubiquitin proteasome system. Interestingly, the cryo-electron microscopy (cryo-EM) structure of the CRBN homodimer reveals a unique mechanism whereby two molecular glues assemble into a helix-like structure and drive ternary complex formation by mimicking a neosubstrate G-loop degron. This CRBN chemical knockout offers a valuable tool to elucidate the molecular mechanism of MGDs, to investigate its endogenous substrates and understand their physiological roles.
- Monte Rosa Therapeutics AG, Basel, Switzerland.
Organizational Affiliation: