a5b3 GABAA Receptor in 1 a5 to 4 b3 stoichiometry in desensitized state
Cowgill, J., Fan, C., Howard, R.J., Lindahl, E.To be published.
Experimental Data Snapshot
Starting Model: in silico
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Green fluorescent protein,Gamma-aminobutyric acid receptor subunit alpha-5 | 679 | Aequorea victoria, Homo sapiens | Mutation(s): 0 Gene Names: GFP, GABRA5 | 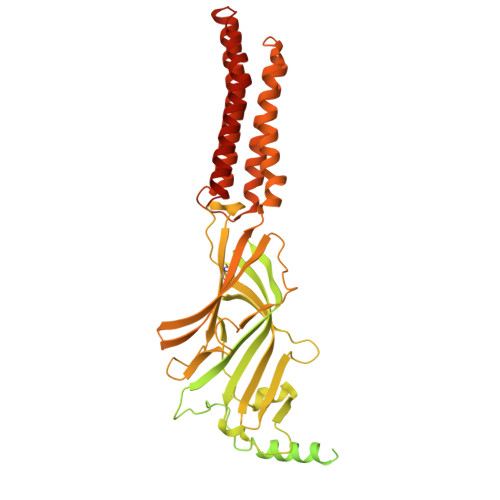 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P31644 (Homo sapiens) Explore P31644 Go to UniProtKB: P31644 | |||||
PHAROS: P31644 GTEx: ENSG00000186297 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P31644 | ||||
Glycosylation | |||||
| Glycosylation Sites: 2 | Go to GlyGen: P31644-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Gamma-aminobutyric acid receptor subunit beta-3,Green fluorescent protein | 623 | Homo sapiens | Mutation(s): 0 Gene Names: GABRB3, GFP | 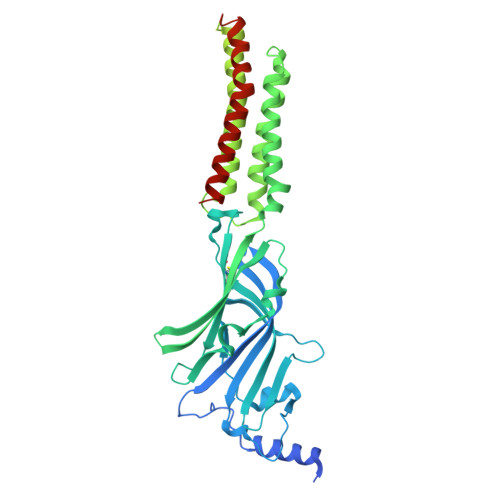 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P28472 (Homo sapiens) Explore P28472 Go to UniProtKB: P28472 | |||||
PHAROS: P28472 GTEx: ENSG00000166206 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P28472 | ||||
Glycosylation | |||||
| Glycosylation Sites: 2 | Go to GlyGen: P28472-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Megabody 25 | F [auth P], G [auth Q], H [auth R] | 541 | Escherichia coli | Mutation(s): 0 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | I [auth F] | 2 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G42666HT GlyCosmos: G42666HT GlyGen: G42666HT | |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | J [auth G], L [auth I], Q [auth N] | 3 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G15407YE GlyCosmos: G15407YE GlyGen: G15407YE | |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | K [auth H], R [auth O] | 6 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G09724ZC GlyCosmos: G09724ZC GlyGen: G09724ZC | |||||
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| beta-D-mannopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | N [auth K], P [auth M] | 3 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G44266EX GlyCosmos: G44266EX GlyGen: G44266EX | |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| EPE Query on EPE | T [auth C], U [auth E] | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID C8 H18 N2 O4 S JKMHFZQWWAIEOD-UHFFFAOYSA-N |  | ||
| ABU (Subject of Investigation/LOI) Query on ABU | S [auth B] | GAMMA-AMINO-BUTANOIC ACID C4 H9 N O2 BTCSSZJGUNDROE-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.21.1_5286 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Swedish Research Council | Sweden | -- |
| European Molecular Biology Organization (EMBO) | European Union | ALTF 542-2021 |