Open-state RyR1 in 0.05% POPC micelles, in complex with a nanobody and FKBP
Li, C., Efremov, R.G.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Currently 9HEO does not have a validation slider image.
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ryanodine receptor 1 | 5,027 | Oryctolagus cuniculus | Mutation(s): 0 Gene Names: RYR1 | 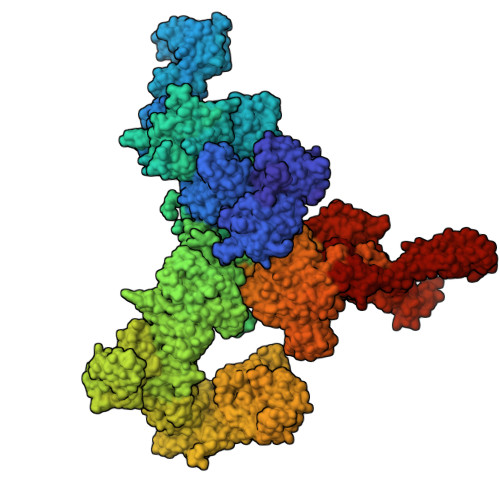 | |
UniProt | |||||
Find proteins for P11716 (Oryctolagus cuniculus) Explore P11716 Go to UniProtKB: P11716 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P11716 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Nanobody 9657 | 126 | Lama glama | Mutation(s): 0 | 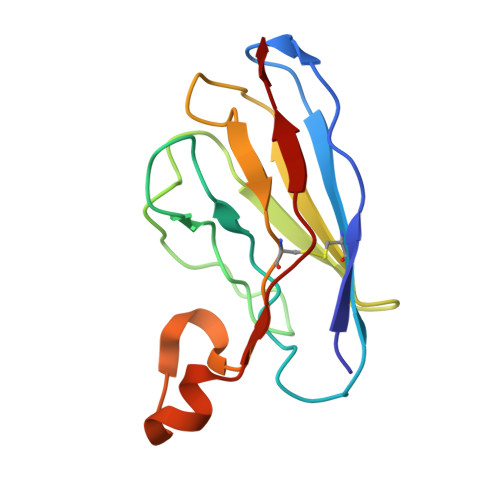 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Peptidyl-prolyl cis-trans isomerase FKBP1B | 107 | Oryctolagus cuniculus | Mutation(s): 0 Gene Names: FKBP1B, FKBP12.6 EC: 5.2.1.8 | 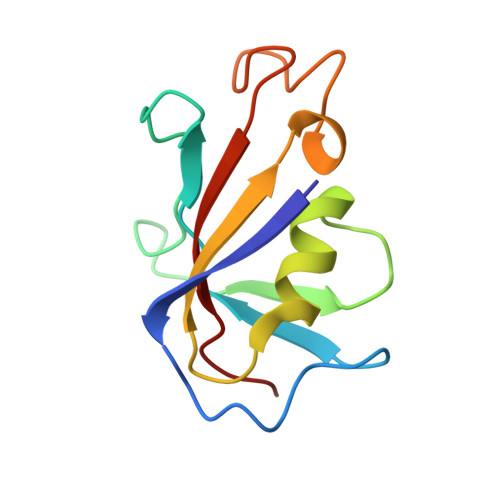 | |
UniProt | |||||
Find proteins for Q8HYX6 (Oryctolagus cuniculus) Explore Q8HYX6 Go to UniProtKB: Q8HYX6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8HYX6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| POV (Subject of Investigation/LOI) Query on POV | AA [auth A] AB [auth C] AC [auth J] BA [auth A] BB [auth C] | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate C42 H82 N O8 P WTJKGGKOPKCXLL-PFDVCBLKSA-N |  | ||
| ATP (Subject of Investigation/LOI) Query on ATP | IB [auth G], IC [auth J], MA [auth C], N [auth A] | ADENOSINE-5'-TRIPHOSPHATE C10 H16 N5 O13 P3 ZKHQWZAMYRWXGA-KQYNXXCUSA-N |  | ||
| CFF (Subject of Investigation/LOI) Query on CFF | JB [auth G], JC [auth J], NA [auth C], O [auth A] | CAFFEINE C8 H10 N4 O2 RYYVLZVUVIJVGH-UHFFFAOYSA-N |  | ||
| ZN Query on ZN | HB [auth G], HC [auth J], LA [auth C], M [auth A] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| CA (Subject of Investigation/LOI) Query on CA | KB [auth G], KC [auth J], OA [auth C], P [auth A] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | cryoSPARC | 4.31 |
Currently 9HEO does not have a validation slider image.
| Funding Organization | Location | Grant Number |
|---|---|---|
| Research Foundation - Flanders (FWO) | Belgium | G0H5916N |
| Research Foundation - Flanders (FWO) | Belgium | G054617N |
| European Research Council (ERC) | European Union | 726436 |