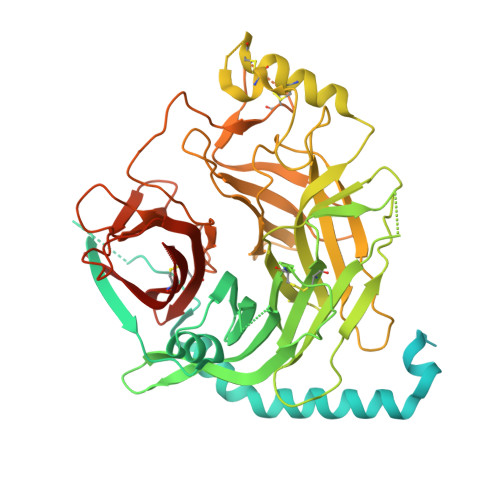
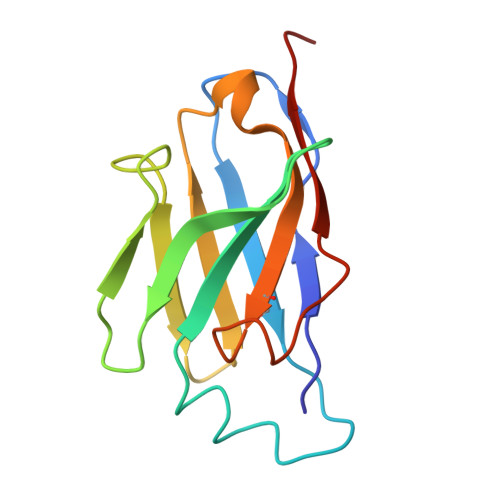
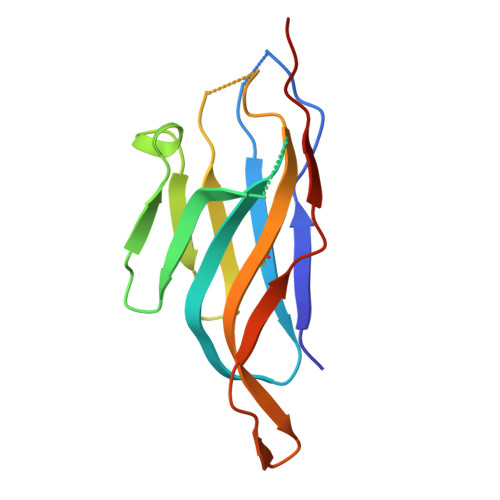
Protection against lethal canine distemper virus infection by a dual epitope-targeting synthetic antibody.
Scherer, M., Djabeur, N., Siering, O., Jeckelmann, J.M., Wyss, M., Cresci, M., Di Palma Subran, M., Riedl, R., Chames, P., Pfaller, C.K., Sawatsky, B., Fotiadis, D., Plattet, P.(2026) Nat Commun 17: 103-103
- PubMed: 41501035
- DOI: https://doi.org/10.1038/s41467-025-67600-z
- Primary Citation of Related Structures:
9HBP - PubMed Abstract:
Despite vaccine availability, the morbilliviruses measles virus and canine distemper virus (CDV) are still causing major health impairments in human and animal populations. Here, we identified two potent, neutralizing single domain antibodies directed against the tetrameric receptor binding (H) protein of CDV. Structural analyses spotlighted two vulnerable sites within the H protein. While the first overlaps with the receptor binding site, the second encompasses amino acid residues of two protomers located at the distal dimeric head interface, which supports distinct mechanisms of neutralization. Upon application of an engineered tetravalent and biparatopic antibody, ferrets were protected at a remarkably low antibody dose (1 mg/kg) administered intra-peritoneally on days 3 and 7 post-exposure of a lethal CDV challenge. Collectively, this study spotlights the power of integrating multiple mechanisms of neutralization in a single format and provides a roadmap to design next-generation therapeutics against morbilliviral infections as well as other infectious pathogens.
- Division of Neurological Sciences, Vetsuisse Faculty, University of Bern, Bern, Switzerland.
Organizational Affiliation: