Crystal structure of Nkp46 in complex with a bicyclic peptide BCY00016132
Pellegrino, S., Bezerra, G.A.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Natural cytotoxicity triggering receptor 1 | 201 | Homo sapiens | Mutation(s): 0 Gene Names: NCR1, LY94 | 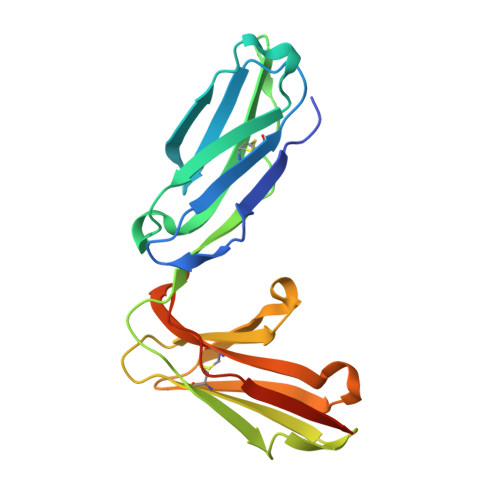 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for O76036 (Homo sapiens) Explore O76036 Go to UniProtKB: O76036 | |||||
PHAROS: O76036 GTEx: ENSG00000189430 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O76036 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Bicyclic peptide BCY00016132 | 16 | synthetic construct | Mutation(s): 0 | 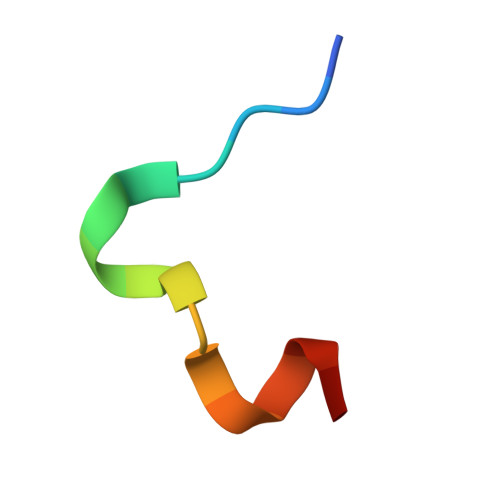 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| LFI Query on LFI | E [auth B] | 1-[3,5-bis(3-bromanylpropanoyl)-1,3,5-triazinan-1-yl]-3-bromanyl-propan-1-one C12 H18 Br3 N3 O3 UYNTZRFVDTZUQI-UHFFFAOYSA-N |  | ||
| DHL Query on DHL | D [auth A] | 2-AMINO-ETHANETHIOL C2 H7 N S UFULAYFCSOUIOV-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | C [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 74.688 | α = 90 |
| b = 74.688 | β = 90 |
| c = 75.307 | γ = 120 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| Aimless | data scaling |
| xia2 | data reduction |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Other private | -- |