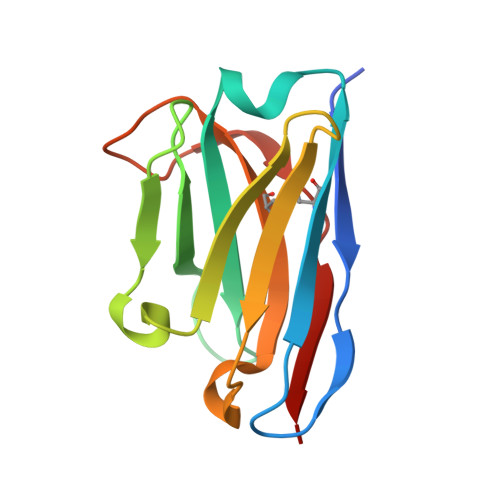
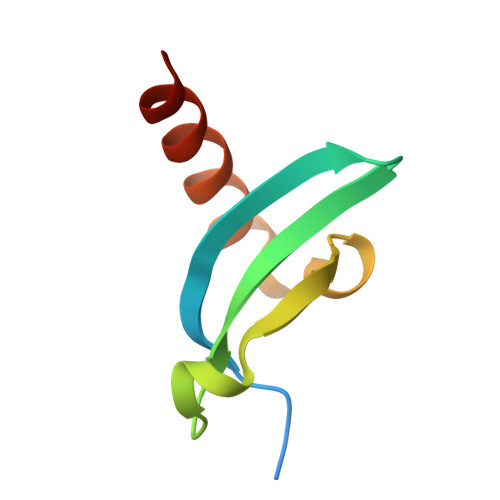
Gluebodies Offer a Route To Improve Crystal Reliability and Diversity through Transferable Nanobody Mutations That Introduce Constitutive Close Contacts.
Ye, M., Makola, M., Richards, M.W., Newman, J.A., Fairhead, M., Burgess, S.G., Wu, Z., Maclean, E., Wright, N.D., Koekemoer, L., Thompson, A., Bezerra, G.A., Yi, G., Li, H., Rangel, V.L., Mamalis, D., Aitkenhead, H., Davis, B.G., Gilbert, R.J.C., Duerr, K.L., Bayliss, R., Gileadi, O., von Delft, F.(2025) ACS Cent Sci 11: 2385-2399
- PubMed: 41473800
- DOI: https://doi.org/10.1021/acscentsci.5c00937
- Primary Citation of Related Structures:
9H77 - PubMed Abstract:
Design of modular, transferable protein assemblies has broad applicability and in structural biology could help with the ever-troublesome crystallization bottleneck, including finding robustly behaved protein crystals for rapidly characterizing ligands or drug candidates or generating multiple polymorphs to illuminate diverse conformations. Nanobodies as crystallization chaperones are well-established but still unreliable, as we show here. Instead, we show an exemplar of how robust crystallization behavior can be engineered by exploring many combinations (>200) of nanobody surface mutations over several iterations. Critically, what needed testing was crystallization and diffraction quality, since target-nanobody binding affinity is decoupled from crystallizability enhancement. Our study yielded multiple polymorphs, all mediated by the same interface, with dramatically improved resolution and diffraction reliability for some mutants; we thus name them 'Gluebodies' (Gbs). We further demonstrate that these Gb mutations do transfer to some other targets, both for achieving robust crystallization in alternative packing forms and for establishing the ability to crystallize a key early stage readout. Since the Gb interface is evidently a favored interaction, it may be broadly applicable for modular assembly; more specifically, this work suggests that Gbs should be routinely attempted for crystallization whenever nanobodies are available.
- Centre for Medicines Discovery, Nuffield Department of Medicine, University of Oxford, Oxford OX3 7FZ, U.K.
Organizational Affiliation: