Structure and function of the yeast amino acid-sensing SEAC-EGOC supercomplex
Tafur, L., Bonadei, L., Zheng, Y., Loewith, R.To be published.
Experimental Data Snapshot
Starting Models: experimental, in silico
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Maintenance of telomere capping protein 5 | A [auth C], I [auth K] | 1,148 | Saccharomyces cerevisiae | Mutation(s): 0 | 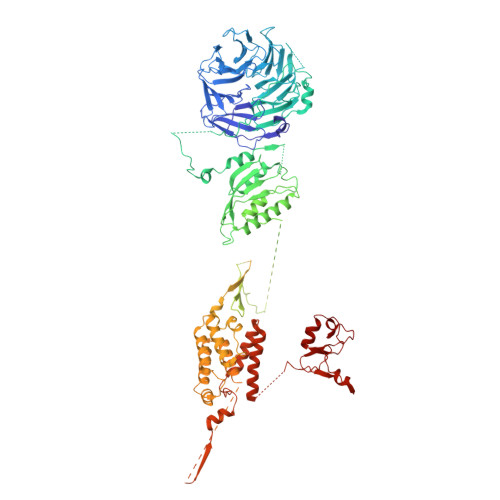 |
UniProt | |||||
Find proteins for Q03897 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore Q03897 Go to UniProtKB: Q03897 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q03897 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein transport protein SEC13 | B [auth H], J [auth P] | 297 | Saccharomyces cerevisiae | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q04491 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore Q04491 Go to UniProtKB: Q04491 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q04491 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Restriction of telomere capping protein 1 | C [auth A], K [auth I] | 1,341 | Saccharomyces cerevisiae | Mutation(s): 0 | 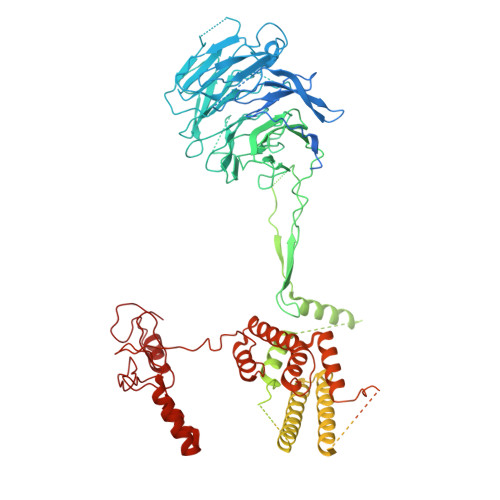 |
UniProt | |||||
Find proteins for Q08281 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore Q08281 Go to UniProtKB: Q08281 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q08281 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Nucleoporin SEH1 | 349 | Saccharomyces cerevisiae | Mutation(s): 0 | 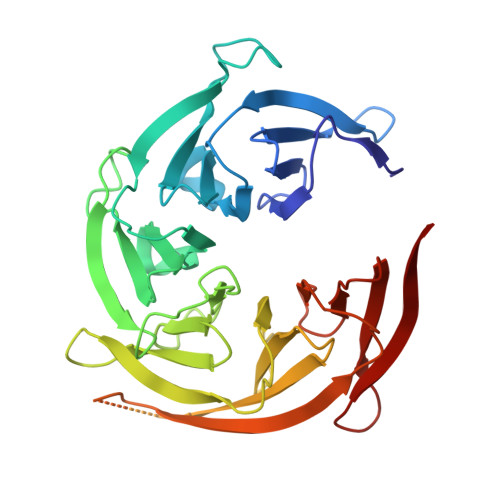 | |
UniProt | |||||
Find proteins for P53011 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P53011 Go to UniProtKB: P53011 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P53011 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| SEH-associated protein 4 | F [auth G], G [auth B], N [auth O], O [auth J] | 1,038 | Saccharomyces cerevisiae | Mutation(s): 0 | 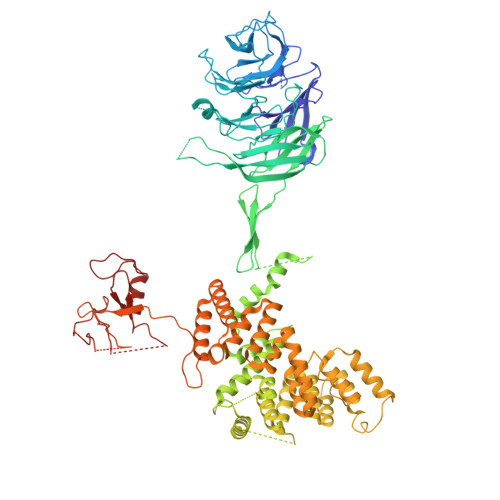 |
UniProt | |||||
Find proteins for P38164 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P38164 Go to UniProtKB: P38164 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P38164 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| GTP-binding protein GTR1 | Q [auth a], Y [auth b] | 310 | Saccharomyces cerevisiae | Mutation(s): 0 Gene Names: GTR1, YML121W, YM7056.05 EC: 3.6.5 |  |
UniProt | |||||
Find proteins for Q00582 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore Q00582 Go to UniProtKB: Q00582 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q00582 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| GTP-binding protein GTR2 | R [auth c], Z [auth d] | 341 | Saccharomyces cerevisiae | Mutation(s): 0 Gene Names: GTR2, YGR163W EC: 3.6.5 |  |
UniProt | |||||
Find proteins for P53290 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P53290 Go to UniProtKB: P53290 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P53290 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Nitrogen permease regulator 2 | AA [auth g], S [auth T] | 615 | Saccharomyces cerevisiae | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P39923 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P39923 Go to UniProtKB: P39923 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P39923 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 9 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Nitrogen permease regulator 3 | BA [auth i], T [auth h] | 1,146 | Saccharomyces cerevisiae | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P38742 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P38742 Go to UniProtKB: P38742 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P38742 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 10 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Vacuolar membrane-associated protein IML1 | CA [auth j], U [auth X] | 1,584 | Saccharomyces cerevisiae | Mutation(s): 0 | 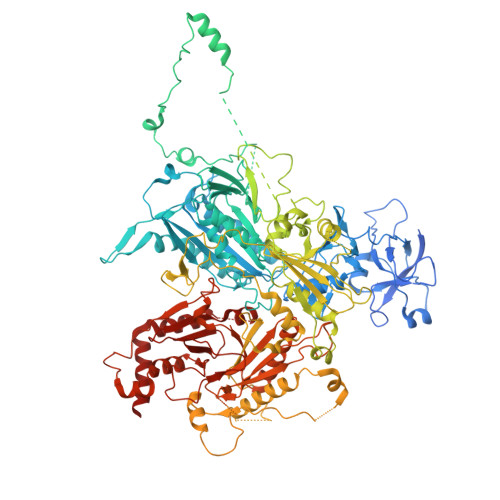 |
UniProt | |||||
Find proteins for P47170 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P47170 Go to UniProtKB: P47170 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P47170 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 11 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein MEH1 | DA [auth S], V [auth R] | 184 | Saccharomyces cerevisiae | Mutation(s): 0 Gene Names: MEH1, EGO1, GSE2, YKR007W, YK106 | 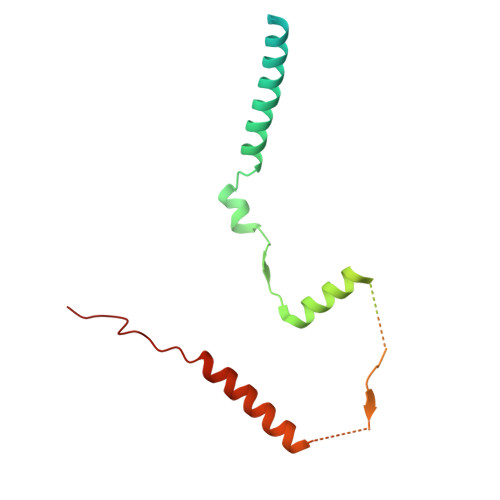 |
UniProt | |||||
Find proteins for Q02205 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore Q02205 Go to UniProtKB: Q02205 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q02205 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 12 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein EGO2 | EA [auth W], W [auth U] | 75 | Saccharomyces cerevisiae | Mutation(s): 0 Gene Names: EGO2, YCR075W-A |  |
UniProt | |||||
Find proteins for Q3E830 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore Q3E830 Go to UniProtKB: Q3E830 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q3E830 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 13 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein SLM4 | FA [auth Z], X [auth Y] | 162 | Saccharomyces cerevisiae | Mutation(s): 0 Gene Names: SLM4, EGO3, GSE1, YBR077C, YBR0723 | 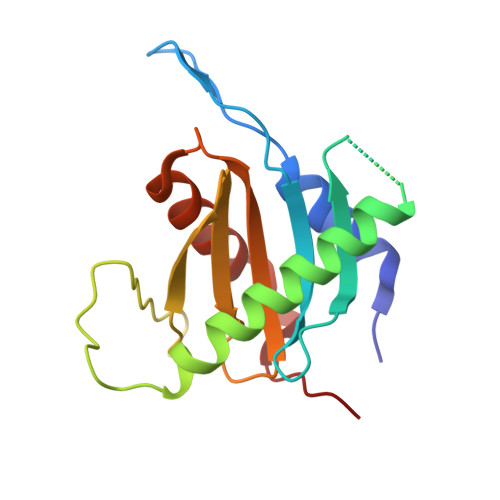 |
UniProt | |||||
Find proteins for P38247 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P38247 Go to UniProtKB: P38247 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P38247 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GDP (Subject of Investigation/LOI) Query on GDP | KB [auth a], LB [auth c], OB [auth b], PB [auth d] | GUANOSINE-5'-DIPHOSPHATE C10 H15 N5 O11 P2 QGWNDRXFNXRZMB-UUOKFMHZSA-N |  | ||
| AF3 (Subject of Investigation/LOI) Query on AF3 | JB [auth a], NB [auth b] | ALUMINUM FLUORIDE Al F3 KLZUFWVZNOTSEM-UHFFFAOYSA-K |  | ||
| ZN (Subject of Investigation/LOI) Query on ZN | AB [auth O] BB [auth O] CB [auth O] DB [auth O] EB [auth J] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| MG (Subject of Investigation/LOI) Query on MG | IB [auth a], MB [auth b] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | cryoSPARC | |
| MODEL REFINEMENT | PHENIX | 1.20 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| European Research Council (ERC) | European Union | AdG TENDO |