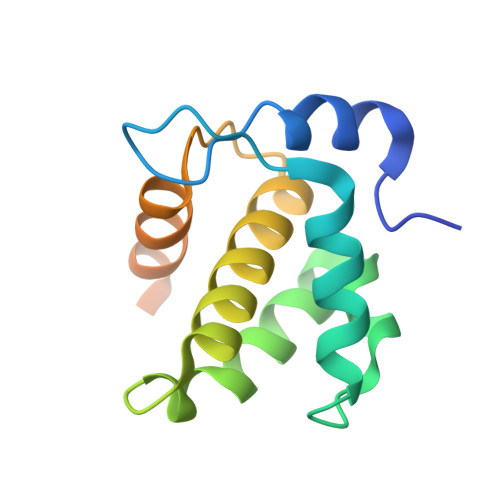
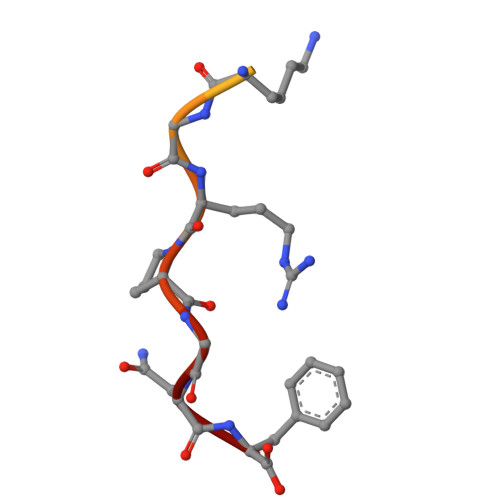
The conserved HIV-1 spacer peptide 2 triggers matrix lattice maturation.
Stacey, J.C.V., Hrebik, D., Nand, E., Shetty, S.D., Qu, K., Boicu, M., Anders-Osswein, M., Uchil, P.D., Dick, R.A., Mothes, W., Krausslich, H.G., Muller, B., Briggs, J.A.G.(2025) Nature 640: 258-264
- PubMed: 40011770
- DOI: https://doi.org/10.1038/s41586-025-08624-9
- Primary Citation of Related Structures:
9H1P - PubMed Abstract:
The virus particles of human immunodeficiency virus type 1 (HIV-1) are released in an immature, non-infectious form. Proteolytic cleavage of the main structural polyprotein Gag into functional domains induces rearrangement into mature, infectious virions. In immature virus particles, the Gag membrane-binding domain, MA, forms a hexameric protein lattice that undergoes structural transition, following cleavage, into a distinct, mature MA lattice 1 . The mechanism of MA lattice maturation is unknown. Here we show that released spacer peptide 2 (SP2), a conserved peptide of unknown function situated about 300 residues downstream of MA, binds MA to induce structural maturation. By high-resolution in-virus structure determination of MA, we show that MA does not bind lipid into a side pocket as previously thought 1 , but instead binds SP2 as an integral part of the protein-protein interfaces that stabilize the mature lattice. Analysis of Gag cleavage site mutants showed that SP2 release is required for MA maturation, and we demonstrate that SP2 is sufficient to induce maturation of purified MA on lipid monolayers in vitro. SP2-triggered MA maturation correlated with faster fusion of virus with target cells. Our results reveal a new, unexpected interaction between two HIV-1 components, provide a high-resolution structure of mature MA, establish the trigger of MA structural maturation and assign function to the SP2 peptide.
- Department of Cell and Virus Structure, Max Planck Institute of Biochemistry, Martinsried, Germany.
Organizational Affiliation: