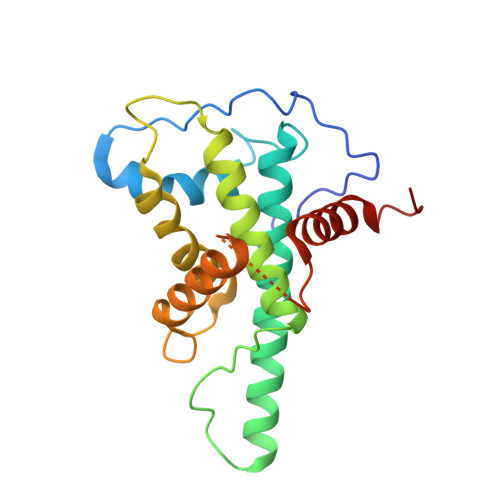
Base-J binding proteins JBP1 and JBP3 have conserved structures of their J-DNA binding domain but drastically different affinities
de Vries, I., Adamopoulos, A., Kazokaite, J., Heidebrecht, T., Fish, A., Celie, P.N.H., Joosten, R.P., Perrakis, A.To be published.