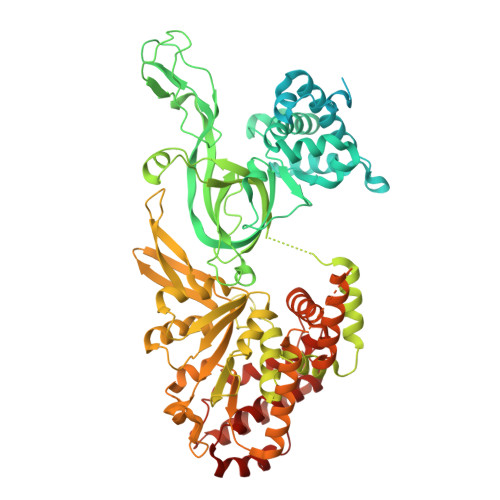
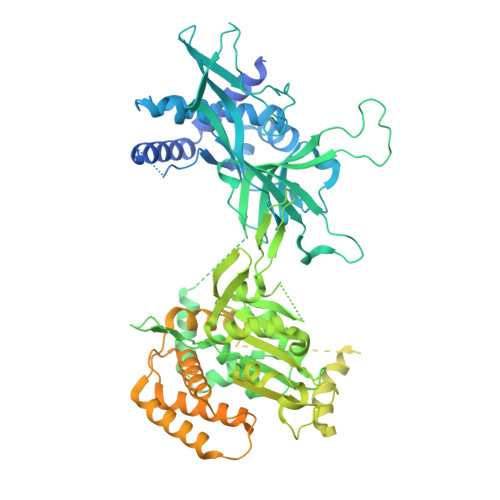
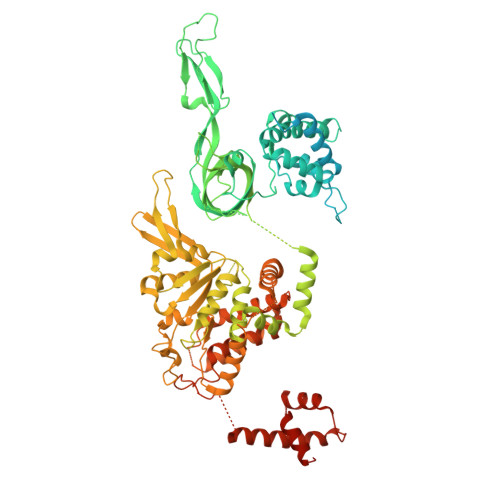
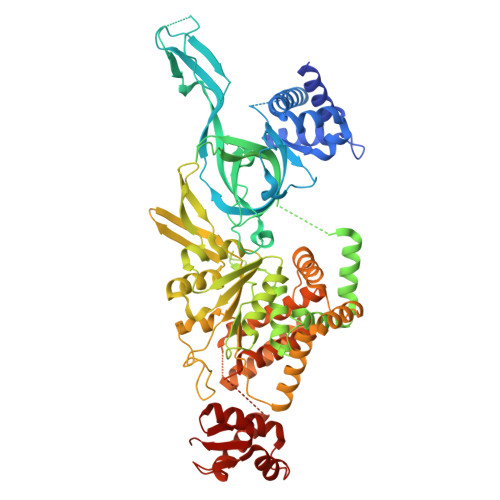
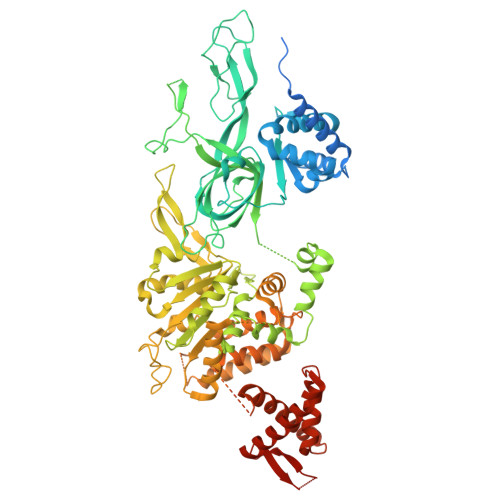
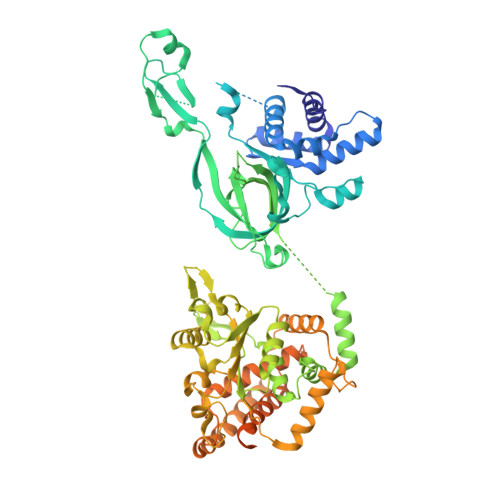
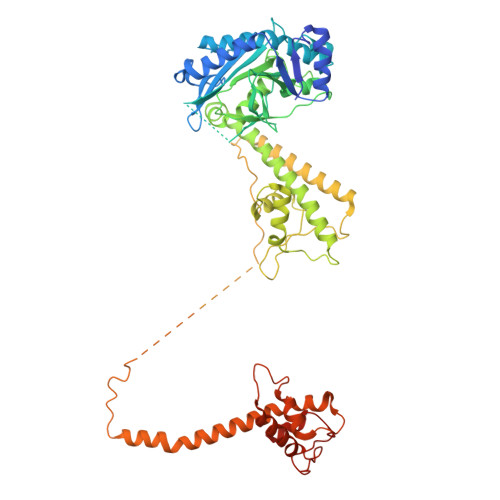
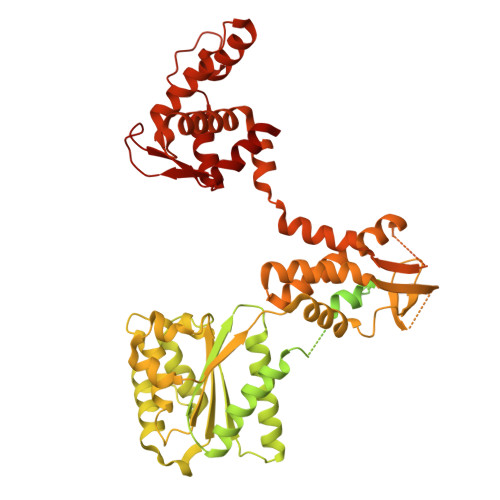
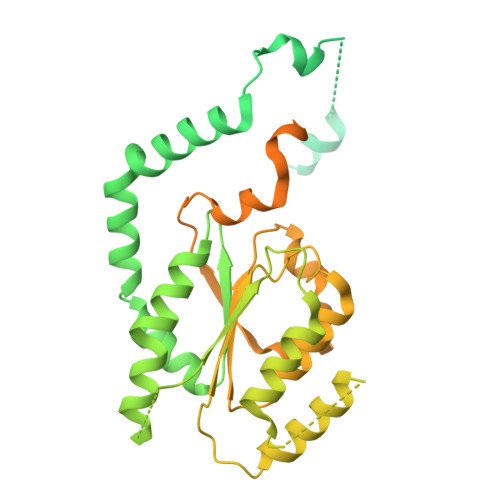
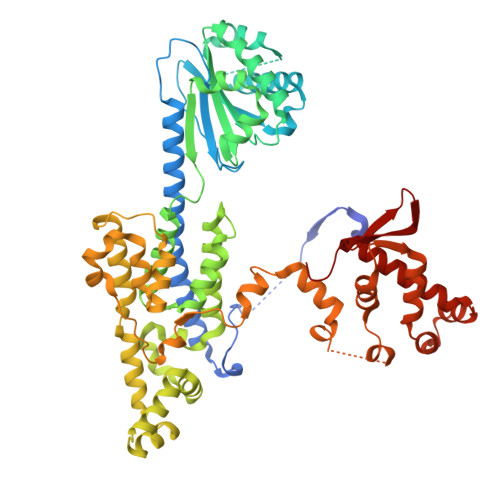
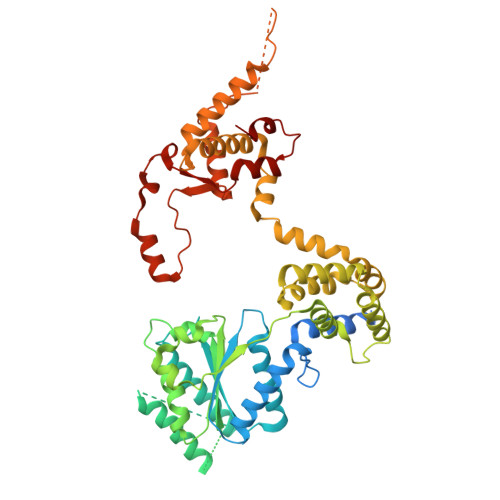
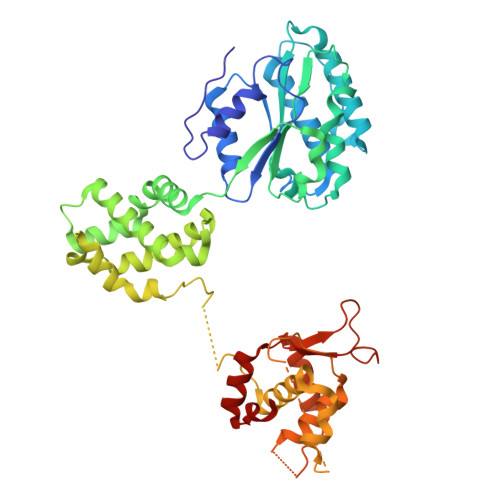
Unidirectional MCM translocation away from ORC drives origin licensing.
Butryn, A., Greiwe, J.F., Costa, A.(2025) Nat Commun 16: 782-782
- PubMed: 39824870
- DOI: https://doi.org/10.1038/s41467-025-56143-y
- Primary Citation of Related Structures:
9GJP, 9GJW, 9GM5 - PubMed Abstract:
The MCM motor of the eukaryotic replicative helicase is loaded as a double hexamer onto DNA by the Origin Recognition Complex (ORC), Cdc6, and Cdt1. ATP binding supports formation of the ORC-Cdc6-Cdt1-MCM (OCCM) helicase-recruitment complex where ORC-Cdc6 and one MCM hexamer form two juxtaposed rings around duplex DNA. ATP hydrolysis by MCM completes MCM loading but the mechanism is unknown. Here, we used cryo-EM to characterise helicase loading with ATPase-dead Arginine Finger variants of the six MCM subunits. We report the structure of two MCM complexes with different DNA grips, stalled as they mature to loaded MCM. The Mcm2 Arginine Finger-variant stabilises DNA binding by Mcm2 away from ORC/Cdc6. The Arginine Finger-variant of the neighbouring Mcm5 subunit stabilises DNA engagement by Mcm5 downstream of the Mcm2 binding site. Cdc6 and Orc1 progressively disengage from ORC as MCM translocates along DNA. We observe that duplex DNA translocation by MCM involves a set of leading-strand contacts by the pre-sensor 1 ATPase hairpins and lagging-strand contacts by the helix-2-insert hairpins. Mutating any of the MCM residues involved impairs high-salt resistant DNA binding in vitro and double-hexamer formation assessed by electron microscopy. Thus, ATPase-powered duplex DNA translocation away from ORC underlies MCM loading.
- Macromolecular Machines Laboratory, The Francis Crick Institute, London, NW1 1AT, UK.
Organizational Affiliation: