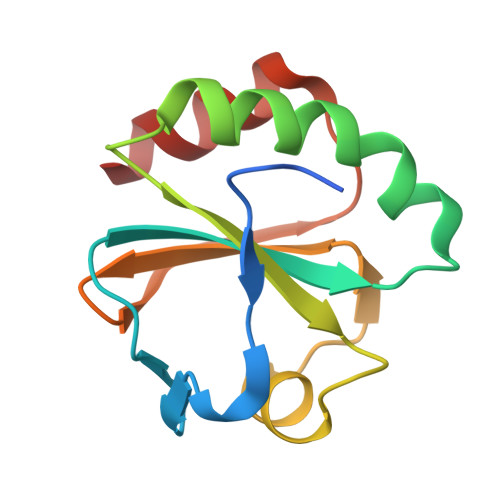
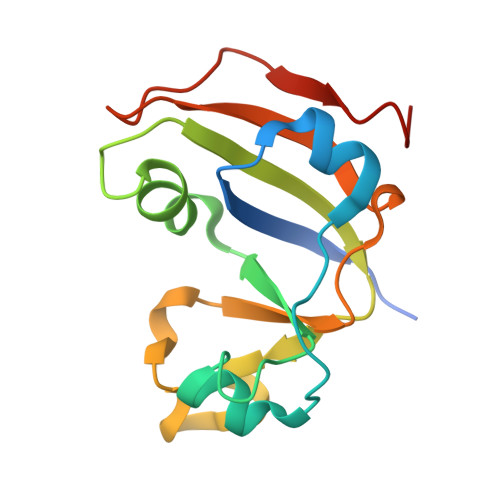
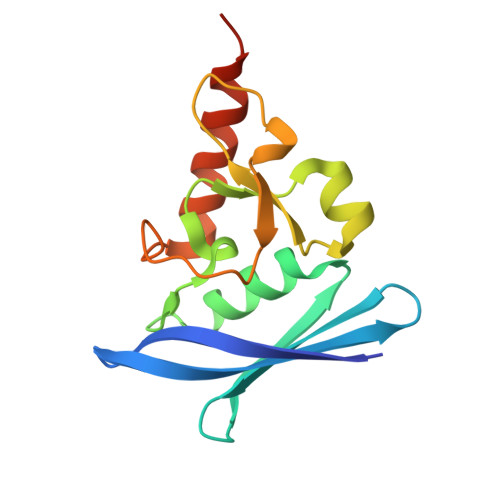
Thioredoxin 1 moonlights as a chaperone for an interbacterial ADP-ribosyltransferase toxin.
Dumont, B., Terradot, L., Cascales, E., Van Melderen, L., Jurenas, D.(2024) Nat Commun 15: 10388-10388
- PubMed: 39613764
- DOI: https://doi.org/10.1038/s41467-024-54892-w
- Primary Citation of Related Structures:
8S2M, 8S2N, 9GCO - PubMed Abstract:
Formation and breakage of disulfide bridges strongly impacts folding and activity of proteins. Thioredoxin 1 (TrxA) is a small, conserved enzyme that reduces disulfide bonds in the bacterial cytosol. In this study, we provide an example of the emergence of a chaperone role for TrxA, which is independent of redox catalysis. We show that the activity of the secreted bacterial ADP-ribosyltransferase (ART) toxin TreX, which does not contain any cysteines, is dependent on TrxA. TreX binds to the reduced form of TrxA via its carboxy-terminal extension to form a soluble and active complex. Structural studies revealed that TreX-like toxins are homologous to Scabin-like ART toxins which possess cysteine residues and form disulfide bridges at the position that superimposes the TrxA binding site in TreX. Our study therefore suggests that thioredoxin 1 evolved alternative functions by maintaining the interaction with cysteine-free substrates.
- Bacterial Genetics and Physiology, Faculté des Sciences, Université Libre de Bruxelles (ULB), Gosselies, Belgium.
Organizational Affiliation: