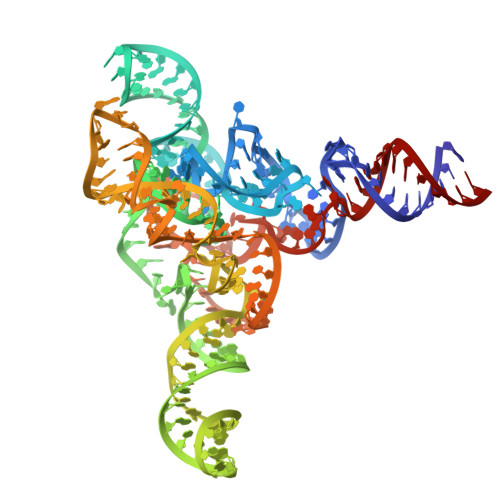
Cryo-EM Structure of raiA ncRNA From Clostridium Reveals a New RNA 3D Fold.
Badepally, N.G., de Moura, T.R., Purta, E., Baulin, E.F., Bujnicki, J.M.(2024) J Mol Biology 436: 168833-168833
- PubMed: 39454748
- DOI: https://doi.org/10.1016/j.jmb.2024.168833
- Primary Citation of Related Structures:
9G7C - PubMed Abstract:
Advancements in genome-wide sequence analysis have led to the discovery of numerous novel bacterial non-coding RNAs (ncRNAs). These ncRNAs have been categorized into various RNA families and classes based on their size, structure, function, and evolutionary relationships. One such ncRNA family, raiA, is notably abundant in the bacterial phyla Firmicutes and Actinobacteria and is remarkably well-conserved across many Gram-positive bacteria. In this study, we integrated cryo-electron microscopy single-particle analysis with computational modeling and biochemical techniques to elucidate the structural characteristics of raiA from Clostridium sp. CAG 138. Our findings reveal the globular 3D fold of raiA, providing valuable structural insights. This analysis paves the way for future investigations into the functional properties of raiA, potentially uncovering new regulatory mechanisms in bacterial ncRNAs.
- Laboratory of Bioinformatics and Protein Engineering, International Institute of Molecular and Cell Biology in Warsaw, ul. Ks. Trojdena 4, 02-109 Warsaw, Poland.
Organizational Affiliation: