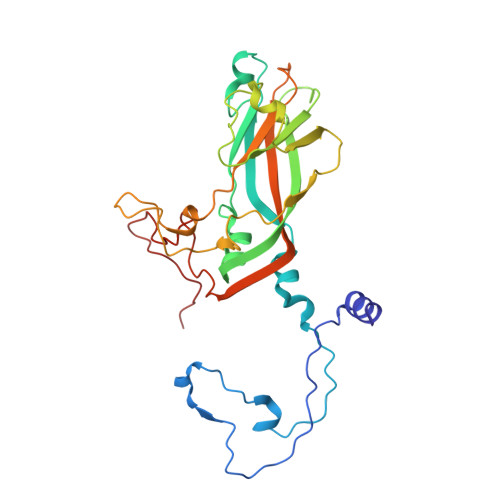
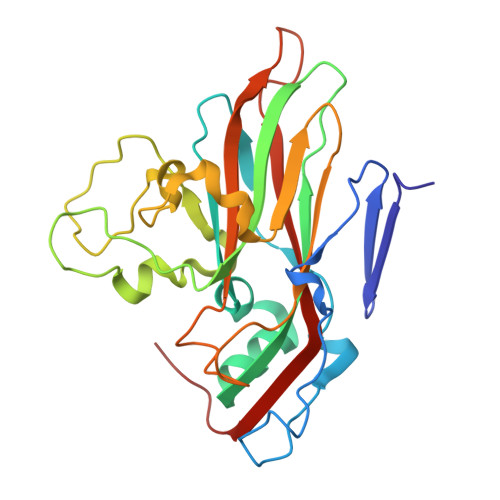
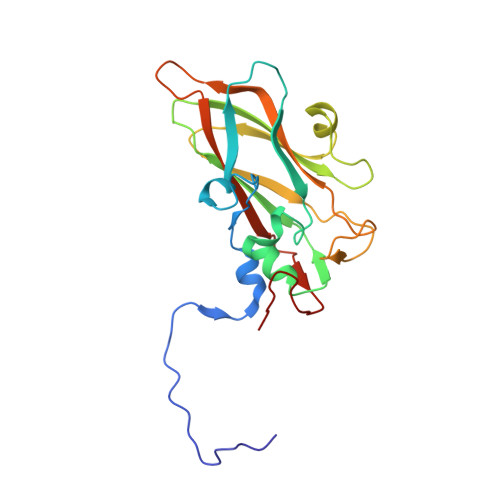
DMSO might impact ligand binding, capsid stability, and RNA interaction in viral preparations.
Wald, J., Goessweiner-Mohr, N., Real-Hohn, A., Blaas, D., Marlovits, T.C.(2024) Sci Rep 14: 30408-30408
- PubMed: 39639094
- DOI: https://doi.org/10.1038/s41598-024-81789-x
- Primary Citation of Related Structures:
9FX1, 9FX9 - PubMed Abstract:
Dimethyl sulfoxide (DMSO) is a widely used solvent in drug research. However, recent studies indicate that even at low concentration DMSO might cause structural changes of proteins and RNA. The pyrazolopyrimidine antiviral OBR-5-340 dissolved in DMSO inhibits rhinovirus-B5 infection yet is inactive against RV-A89. This is consistent with our structural observation that OBR-5-340 is only visible at the pocket factor site in rhinovirus-B5 and not in RV-A89, where the hydrophobic pocket is collapsed. Here, we analyze the impact of DMSO in RV-A89 by high-resolution cryo-electron microscopy. Our 1.76 Å cryo-EM reconstruction of RV-A89 in plain buffer, without DMSO, reveals that the pocket-factor binding site is occupied by myristate and that the previously observed local heterogeneity at protein-RNA interfaces is absent. These findings suggest that DMSO elutes the pocket factor, leading to a collapse of the hydrophobic pocket of RV-A89. Consequently, the conformational heterogeneity observed at the RNA-protein interface in the presence of DMSO likely results from increased capsid flexibility due to the absence of the pocket factor and DMSO-induced affinity modifications. This local asymmetry may promote a directional release of the RNA genome during infection.
- Institute of Microbial and Molecular Sciences, University Medical Center Hamburg-Eppendorf, Hamburg, Germany. jiri.wald@cssb-hamburg.de.
Organizational Affiliation: