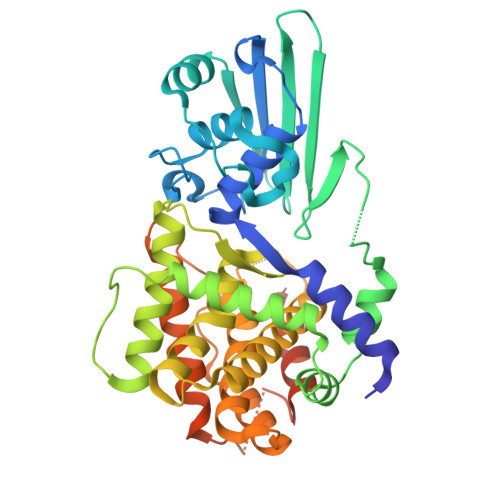
Evolution, structure and function of L-cysteine desulfidase, an enzyme involved in sulfur metabolism in the methanogenic archeon Methanococcus maripaludis.
Gervason, S., Zecchin, P., Shelton, E.B., He, N., Pecqueur, L., Garcia, P.S., Akinyemi, T., Touati, N., Bimai, O., Velours, C., Ravanat, J.L., Faivre, B., Whitman, W.B., Fontecave, M., Golinelli-Pimpaneau, B.(2025) Commun Biol 8: 1667-1667
- PubMed: 41291062
- DOI: https://doi.org/10.1038/s42003-025-09053-0
- Primary Citation of Related Structures:
9FSL - PubMed Abstract:
The biosynthesis of sulfur-containing molecules, which play essential roles in cell metabolism, often relies on enzymes that mobilize sulfur from cysteine. The function of such enzyme, L-cysteine desulfidase CyuA, which catalyzes L-cysteine decomposition to pyruvate, ammonia, and hydrogen sulfide, remains incompletely understood. Here, we used phylogenetic, genetic, biochemical, spectroscopic, and structural approaches to connect molecular structure to cellular physiology and evolutionary history and elucidate CyuA's role in sulfur metabolism. We found that Methanococcales and several other archaeal lineages acquired CyuA via horizontal gene transfer from bacteria. In Methanococcus maripaludis, CyuA (MmCyuA) stimulates growth in sulfide-rich conditions and enables slow growth with cysteine as the sole sulfur source. Crystallographic and biochemical data reveal that MmCyuA binds a [4Fe-4S] cluster coordinated by three conserved cysteines; the fourth ligand is a nonconserved cysteine in the wild-type enzyme but is replaced by glycerol or ethylene glycol in a variant. These results enabled modeling of the enzyme-substrate complex, allowing us to propose a detailed mechanism for L-cysteine desulfuration by CyuA, potentially involving a transient [4Fe-5S] species to transfer sulfur from cysteine to various [4Fe-4S]-dependent tRNA sulfuration enzymes. These findings advance understanding of sulfur activation and trafficking related to biosynthetic pathways leading to sulfur-containing compounds.
- Laboratoire de Chimie des Processus Biologiques, UMR 8229 CNRS, Collège de France, Sorbonne Université, Paris, France.
Organizational Affiliation: