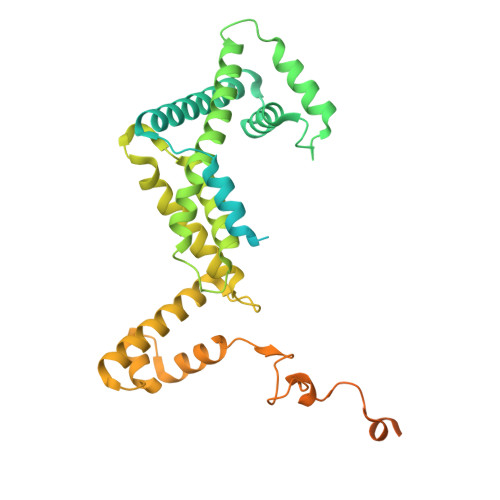
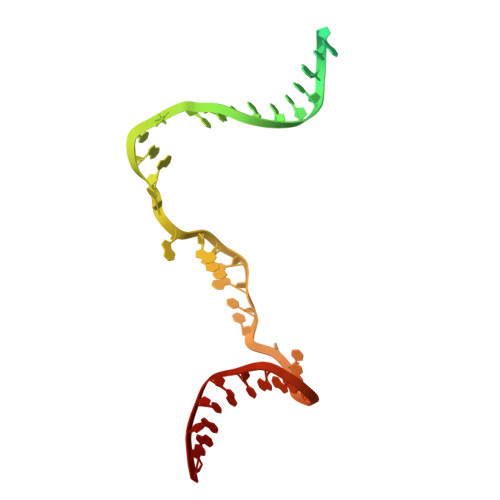
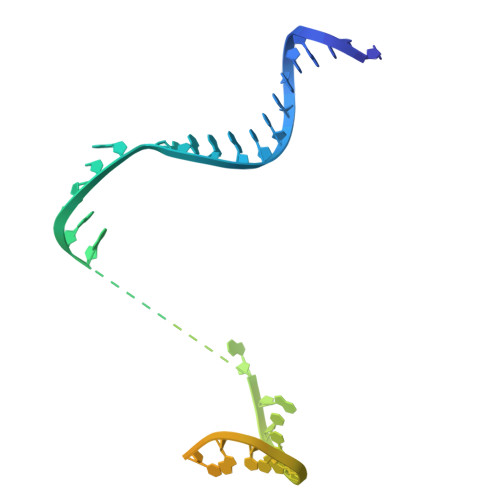
Molecular insight into 5' RNA capping with Np n Ns by bacterial RNA polymerase.
Serianni, V.M., Skerlova, J., Dubankova, A.K., Skriba, A., Svachova, H., Vuckova, T., Filimonenko, A., Fabry, M., Rezacova, P., Kouba, T., Cahova, H.(2026) Nat Chem Biol
- PubMed: 41513851
- DOI: https://doi.org/10.1038/s41589-025-02134-5
- Primary Citation of Related Structures:
9FO6, 9FOG, 9FOK, 9FP3, 9FRJ, 9R75 - PubMed Abstract:
RNA capped with dinucleoside polyphosphates has been discovered in bacteria and eukaryotes only recently. The likely mechanism of this specific capping involves direct incorporation of dinucleoside polyphosphates by RNA polymerase as noncanonical initiating nucleotides. However, how these compounds bind into the active site of RNA polymerase during transcription initiation is unknown. Here, we explored transcription initiation in vitro, using a series of DNA templates in combination with dinucleoside polyphosphates and model RNA polymerase from Thermus thermophilus. We observed that the transcription start site can vary on the basis of the compatibility of the specific template and dinucleoside polyphosphate. Cryo-electron microscopy structures of transcription initiation complexes with dinucleoside polyphosphates revealed that both nucleobase moieties can pair with the DNA template. The first encoded nucleotide pairs in a canonical Watson-Crick manner, whereas the second nucleobase pairs noncanonically in a reverse Watson-Crick manner. Our work provides a structural explanation of how dinucleoside polyphosphates initiate RNA transcription.
- Institute of Organic Chemistry and Biochemistry of the CAS, Flemingovo náměstí 2, Prague, Czechia.
Organizational Affiliation: