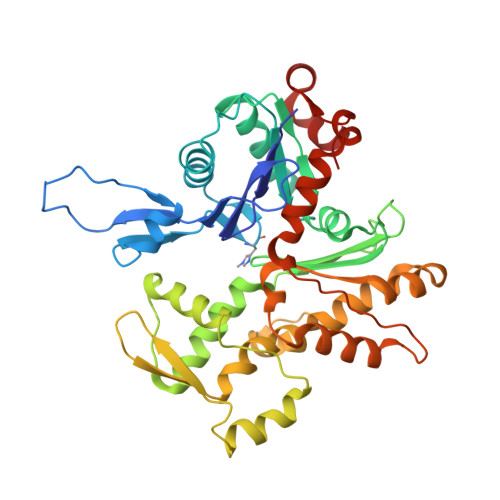
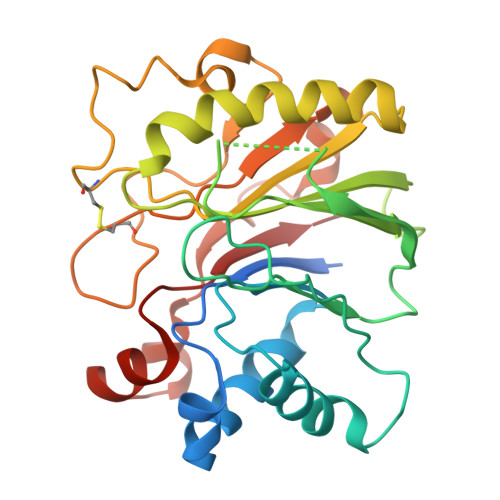
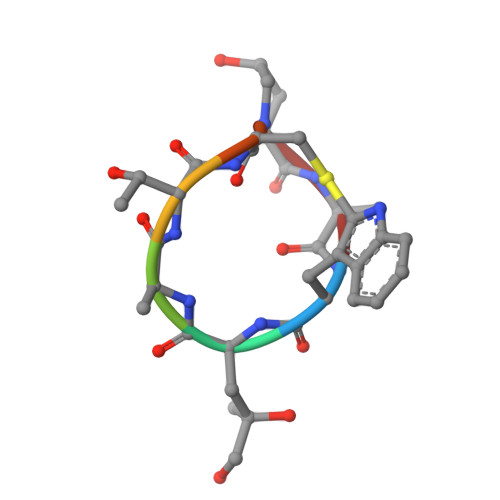
Phalloidin and DNase I-bound F-actin pointed end structures reveal principles of filament stabilization and disassembly.
Boiero Sanders, M., Oosterheert, W., Hofnagel, O., Bieling, P., Raunser, S.(2024) Nat Commun 15: 7969-7969
- PubMed: 39261469
- DOI: https://doi.org/10.1038/s41467-024-52251-3
- Primary Citation of Related Structures:
9FJM, 9FJO, 9FJU, 9FJY - PubMed Abstract:
Actin filament turnover involves subunits binding to and dissociating from the filament ends, with the pointed end being the primary site of filament disassembly. Several molecules modulate filament turnover, but the underlying mechanisms remain incompletely understood. Here, we present three cryo-EM structures of the F-actin pointed end in the presence and absence of phalloidin or DNase I. The two terminal subunits at the undecorated pointed end adopt a twisted conformation. Phalloidin can still bind and bridge these subunits, inducing a conformational shift to a flattened, F-actin-like state. This explains how phalloidin prevents depolymerization at the pointed end. Interestingly, two DNase I molecules simultaneously bind to the phalloidin-stabilized pointed end. In the absence of phalloidin, DNase I binding would disrupt the terminal actin subunit packing, resulting in filament disassembly. Our findings uncover molecular principles of pointed end regulation and provide structural insights into the kinetic asymmetry between the actin filament ends.
- Department of Structural Biochemistry, Max Planck Institute of Molecular Physiology, 44227, Dortmund, Germany.
Organizational Affiliation: