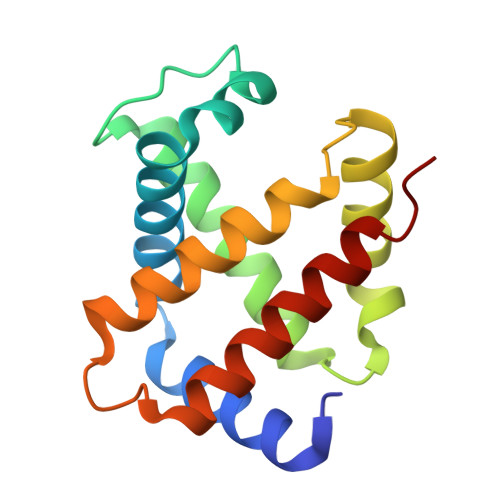
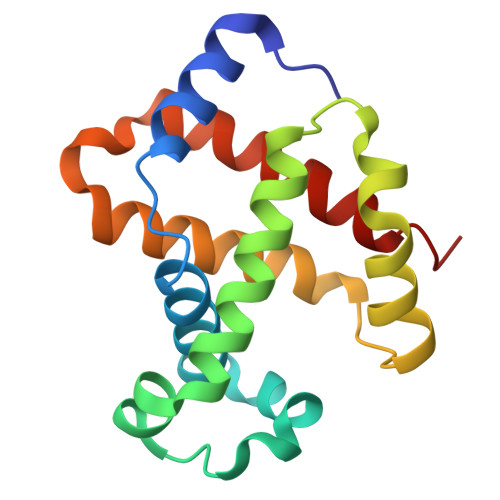
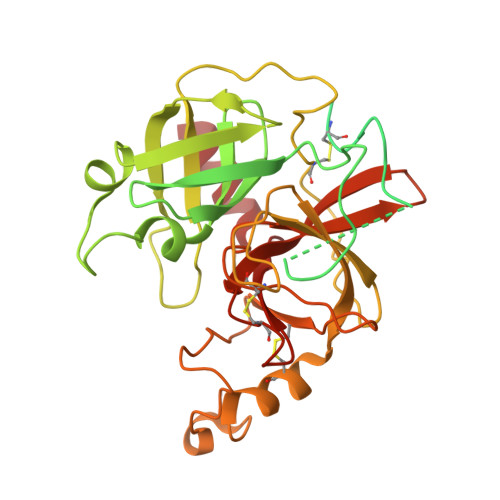
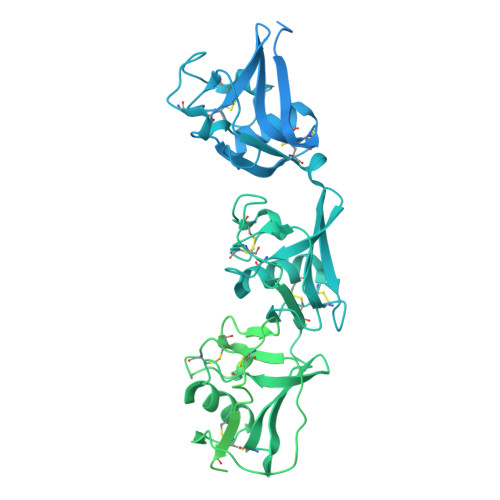
The Cryo-EM structure of human CD163 bound to haptoglobin-hemoglobin reveals molecular mechanisms of hemoglobin scavenging.
Etzerodt, A., Mikkelsen, J.H., Torvund-Jensen, M., Hennig, D., Boesen, T., Graversen, J.H., Moestrup, S.K., Kollman, J.M., Andersen, C.B.F.(2024) Nat Commun 15: 10871-10871
- PubMed: 39738064
- DOI: https://doi.org/10.1038/s41467-024-55171-4
- Primary Citation of Related Structures:
9FHB, 9FMU, 9FNM, 9FNO - PubMed Abstract:
CD163, a macrophage-specific receptor, plays a critical role in scavenging hemoglobin released during hemolysis, protecting against oxidative effects of heme iron. In the bloodstream, hemoglobin is bound by haptoglobin, leading to its immediate endocytosis by CD163. While haptoglobin's structure and function are well understood, CD163's structure and its interaction with the haptoglobin-hemoglobin complex have remained elusive. Here, we present the cryo-electron microscopy structure of the entire extracellular domain of human CD163 in complex with haptoglobin-hemoglobin. The structure reveals that CD163 assembles into trimers (and to some extent dimers), binding haptoglobin-hemoglobin in their center. Key acidic residues in CD163 interact with lysine residues from both haptoglobin and hemoglobin. Calcium-binding sites located near the haptoglobin-hemoglobin interface in CD163 provide explanation for the calcium dependence of the interaction. Furthermore, we show that the interaction facilitating CD163 oligomerization mimics ligand binding and is also calcium dependent. This structural insight into CD163 advances our understanding of its role in hemoglobin scavenging as well as its broader relevance to structurally related scavenger receptors.
- Department of Biomedicine, Aarhus University, 8000, Aarhus C, Denmark.
Organizational Affiliation: