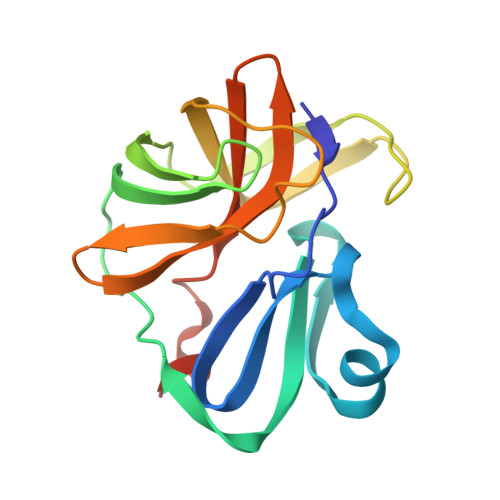
Crystal structure of Enterovirus 71 2A protease mutant C110A containing VP1-2A junction in the active site
Ni, X., Koekemoer, L., Williams, E.P., Wang, S., Wright, N.D., Godoy, A.S., Aschenbrenner, J.C., Balcomb, B.H., Lithgo, R.M., Marples, P.G., Fairhead, M., Thompson, W., Kirkegaard, K., Fearon, D., Walsh, M.A., von Delft, F.To be published.