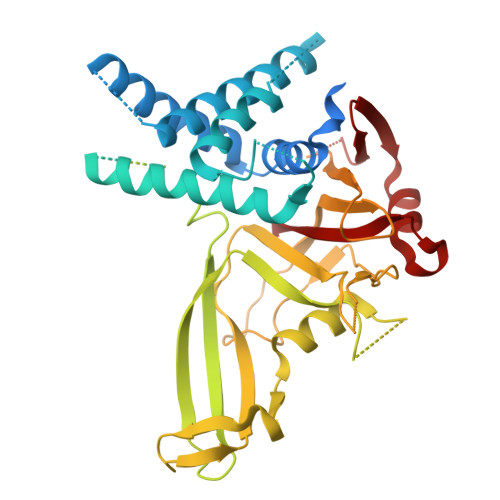
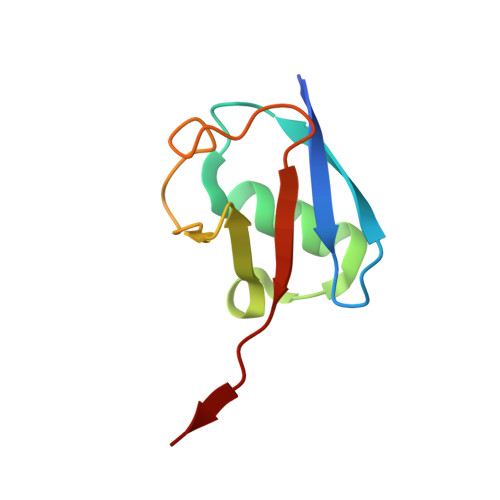
Structural and Biochemical Insights into the Mechanism of Action of the Clinical USP1 Inhibitor, KSQ-4279.
Rennie, M.L., Gundogdu, M., Arkinson, C., Liness, S., Frame, S., Walden, H.(2024) J Med Chem 67: 15557-15568
- PubMed: 39190802
- DOI: https://doi.org/10.1021/acs.jmedchem.4c01184
- Primary Citation of Related Structures:
9FCI, 9FCJ - PubMed Abstract:
DNA damage triggers cell signaling cascades that mediate repair. This signaling is frequently dysregulated in cancers. The proteins that mediate this signaling are potential targets for therapeutic intervention. Ubiquitin-specific protease 1 (USP1) is one such target, with small-molecule inhibitors already in clinical trials. Here, we use biochemical assays and cryo-electron microscopy (cryo-EM) to study the clinical USP1 inhibitor, KSQ-4279 (RO7623066), and compare this to the well-established tool compound, ML323. We find that KSQ-4279 binds to the same cryptic site of USP1 as ML323 but disrupts the protein structure in subtly different ways. Inhibitor binding drives a substantial increase in thermal stability of USP1, which may be mediated through the inhibitors filling a hydrophobic tunnel-like pocket in USP1. Our results contribute to the understanding of the mechanism of action of USP1 inhibitors at the molecular level.
- School of Molecular Biosciences, College of Medical Veterinary and Life Sciences, University of Glasgow, Glasgow G12 8QQ, U.K.
Organizational Affiliation: