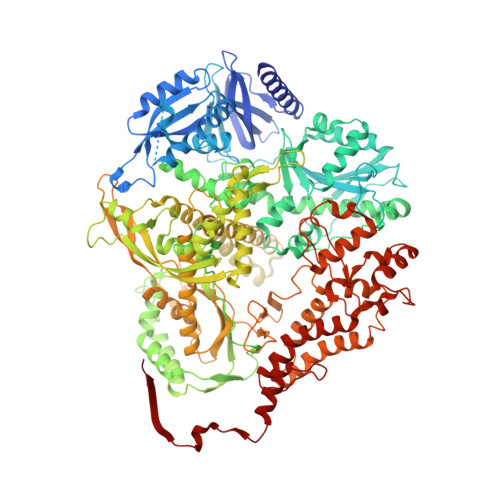
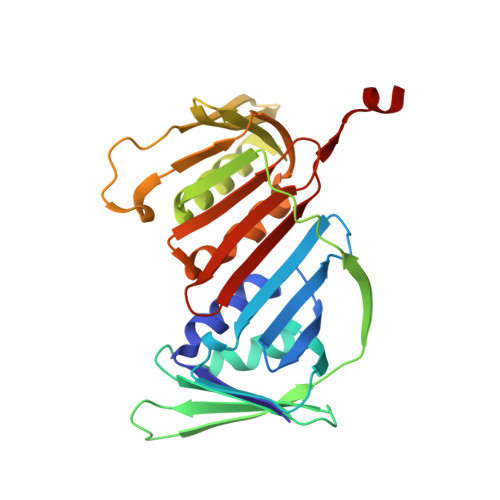
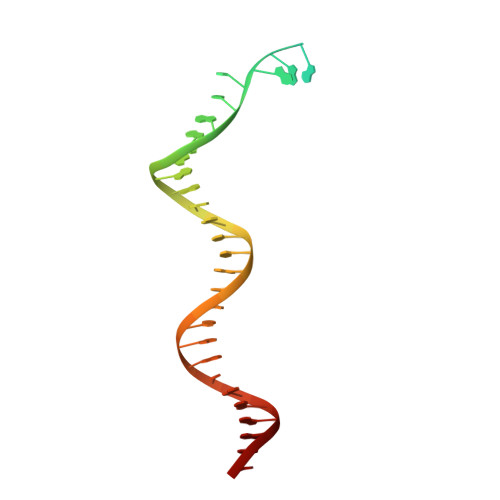
Structural basis for processive daughter-strand synthesis and proofreading by the human leading-strand DNA polymerase Pol epsilon.
Roske, J.J., Yeeles, J.T.P.(2024) Nat Struct Mol Biol 31: 1921-1931
- PubMed: 39112807
- DOI: https://doi.org/10.1038/s41594-024-01370-y
- Primary Citation of Related Structures:
9F6D, 9F6E, 9F6F, 9F6I, 9F6J, 9F6K, 9F6L - PubMed Abstract:
During chromosome replication, the nascent leading strand is synthesized by DNA polymerase epsilon (Pol ε), which associates with the sliding clamp processivity factor proliferating cell nuclear antigen (PCNA) to form a processive holoenzyme. For high-fidelity DNA synthesis, Pol ε relies on nucleotide selectivity and its proofreading ability to detect and excise a misincorporated nucleotide. Here, we present cryo-electron microscopy (cryo-EM) structures of human Pol ε in complex with PCNA, DNA and an incoming nucleotide, revealing how Pol ε associates with PCNA through its PCNA-interacting peptide box and additional unique features of its catalytic domain. Furthermore, by solving a series of cryo-EM structures of Pol ε at a mismatch-containing DNA, we elucidate how Pol ε senses and edits a misincorporated nucleotide. Our structures delineate steps along an intramolecular switching mechanism between polymerase and exonuclease activities, providing the basis for a proofreading mechanism in B-family replicative polymerases.
- MRC Laboratory of Molecular Biology, Cambridge, UK.
Organizational Affiliation: