S. cerevisiae pH nine-sensitive protein 1 is not a choline transporter.
Driller, J.H., Pedersen, B.P.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein PNS1 | 572 | Saccharomyces cerevisiae | Mutation(s): 0 Gene Names: PNS1, YOR161C, O3568 Membrane Entity: Yes | 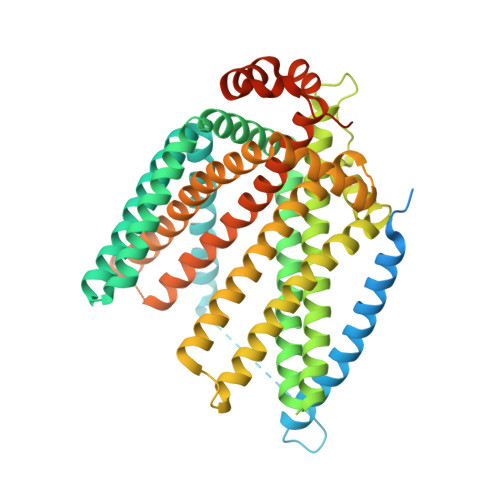 | |
UniProt | |||||
Find proteins for Q12412 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore Q12412 Go to UniProtKB: Q12412 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q12412 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 67.449 | α = 90 |
| b = 91.262 | β = 90 |
| c = 108.372 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| Aimless | data scaling |
| XDS | data reduction |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Lundbeckfonden | Denmark | R380-2021-1207 |
| H2020 Marie Curie Actions of the European Commission | European Union | 890822 |