Development of a first-in-class antibody and a specific assay for alpha-1,6-fucosylated prostate-specific antigen.
Halldorsson, S., Hillringhaus, L., Hojer, C., Muranyi, A., Schraeml, M., Lange, M.S., Tabares, G.(2024) Sci Rep 14: 16512-16512
- PubMed: 39020051
- DOI: https://doi.org/10.1038/s41598-024-67545-1
- Primary Citation of Related Structures:
9F18, 9F1I - PubMed Abstract:
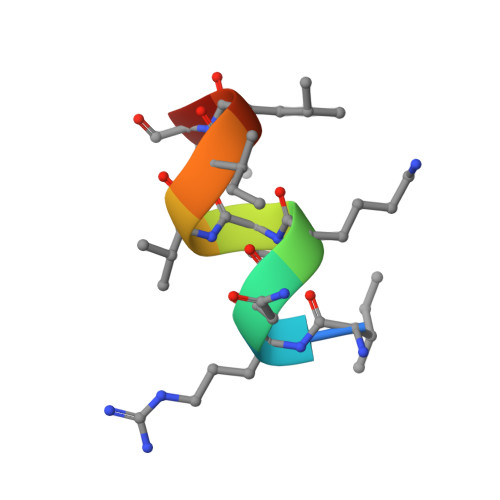
Prostate-specific antigen (PSA) levels are widely used to screen for prostate cancer, yet the test has poor sensitivity, specificity and predictive value, which leads to overdiagnosis and overtreatment. Alterations in the glycosylation status of PSA, including fucosylation, may offer scope for an improved biomarker. We sought to generate a monoclonal antibody (mAb) targeting α-1,6-fucosylated PSA (fuc-PSA) and to develop a tissue-based immunological assay for fuc-PSA detection. Immunogens representing fuc-PSA were used for immunisation and resultant mAbs were extensively characterised. The mAbs reacted specifically with fuc-PSA-specific glycopeptide, but not with aglycosylated PSA or glycan without the PSA peptide. Reactivity was confirmed using high-throughput surface plasmon resonance spectroscopy. X-ray crystallography investigations showed that the mAbs bound to an α-helical form of the peptide, whereas the native PSA epitope is linear. Protein unfolding was required for detection of fuc-PSA in patient samples. Peptide inhibition of fuc-PSA mAbs was observed with positive screening reagents, and target epitope specificity was observed in formalin-fixed, paraffin-embedded tissue samples. This research introduces a well-characterised, first-in-class antibody targeting fuc-PSA and presents the first crystal structure of an antibody demonstrating glycosylation-specific binding to a peptide.
Organizational Affiliation:
Pharma Research and Early Development, F. Hoffmann-La Roche AG, Basel, Switzerland.