Monomeric rhubarb fluorescent protein
Aksakal, O., Rizkallah, P.J., Jones, D.D.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| mRhubarb720 fluorescent protein | 304 | Rhodopseudomonas palustris | Mutation(s): 7 Gene Names: iRFP | 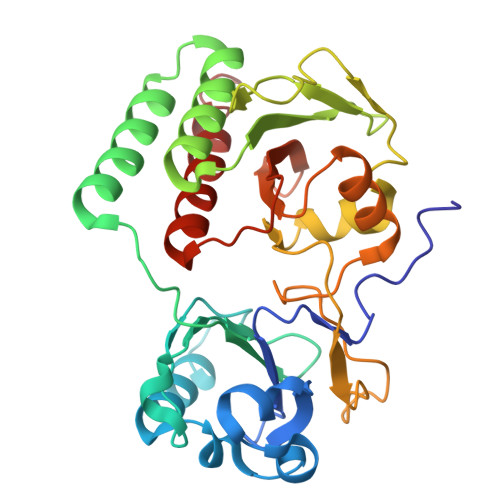 | |
UniProt | |||||
Find proteins for G1FNL7 (Rhodopseudomonas palustris) Explore G1FNL7 Go to UniProtKB: G1FNL7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | G1FNL7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| LBV (Subject of Investigation/LOI) Query on LBV | B [auth A] | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium
-2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3-
yl]propanoic acid C33 H37 N4 O6 DKMLMZVDTGOEGU-ISEYCTJISA-O |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 58.845 | α = 90 |
| b = 58.845 | β = 90 |
| c = 185.883 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| Aimless | data scaling |
| xia2 | data reduction |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Engineering and Physical Sciences Research Council | United Kingdom | -- |