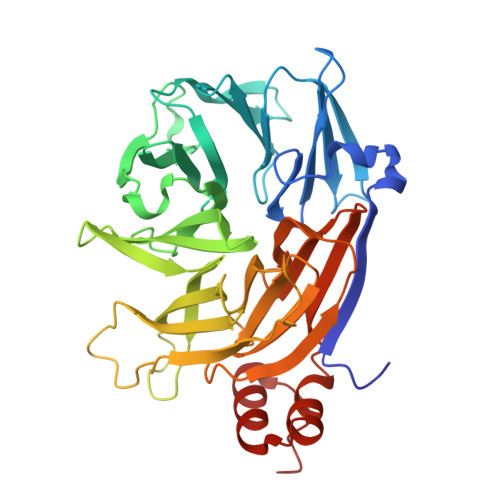
Subtleties in Clathrin heavy chain binding boxes provide selectivity among adaptor proteins of budding yeast.
Defelipe, L.A., Veith, K., Burastero, O., Kupriianova, T., Bento, I., Skruzny, M., Kolbel, K., Uetrecht, C., Thuenauer, R., Garcia-Alai, M.M.(2024) Nat Commun 15: 9655-9655
- PubMed: 39511183
- DOI: https://doi.org/10.1038/s41467-024-54037-z
- Primary Citation of Related Structures:
9EX5, 9EXF, 9EXG, 9EXT, 9EYT - PubMed Abstract:
Clathrin forms a triskelion, or three-legged, network that regulates cellular processes by facilitating cargo internalization and trafficking in eukaryotes. Its N-terminal domain is crucial for interacting with adaptor proteins, which link clathrin to the membrane and engage with specific cargo. The N-terminal domain contains up to four adaptor-binding sites, though their role in preferential occupancy by adaptor proteins remains unclear. In this study, we examine the binding hierarchy of adaptors for clathrin, using integrative biophysical and structural approaches, along with in vivo functional experiments. We find that yeast epsin Ent5 has the highest affinity for clathrin, highlighting its key role in cellular trafficking. Epsins Ent1 and Ent2, crucial for endocytosis but thought to have redundant functions, show distinct binding patterns. Ent1 exhibits stronger interactions with clathrin than Ent2, suggesting a functional divergence toward actin binding. These results offer molecular insights into adaptor protein selectivity, suggesting they competitively bind clathrin while also targeting three different clathrin sites.
- European Molecular Biology Laboratory - Hamburg Unit, Hamburg, Germany.
Organizational Affiliation: