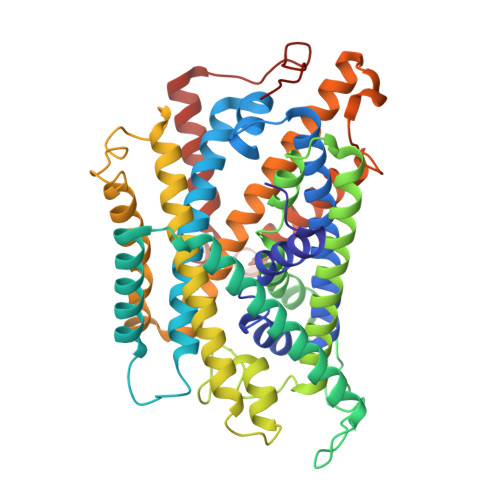
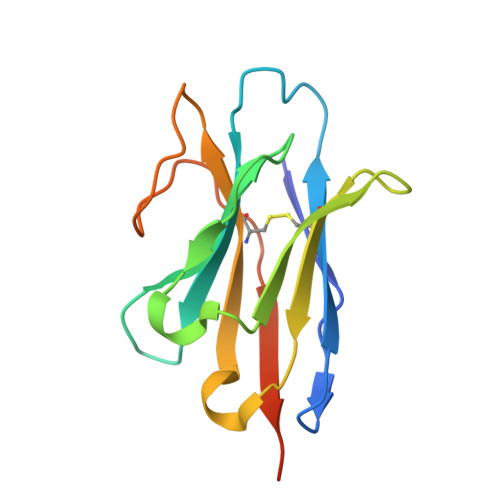
Structural basis of specific lysine transport by Pseudomonas aeruginosa permease LysP.
Bicer, D., Matsuoka, R., Moumbock, A.F.A., Sukumar, P., Suades, A., Cheruvara, H., Quigley, A., Drew, D., Pardon, E., Steyaert, J., Henderson, P.J.F., Caffrey, M., Griese, J.J., Nji, E.(2025) Nat Commun 17: 37-37
- PubMed: 41345107
- DOI: https://doi.org/10.1038/s41467-025-66618-7
- Primary Citation of Related Structures:
9EYD - PubMed Abstract:
Under conditions of extreme acidity, the lysine-specific permease, LysP, not only mediates the import of L-lysine it also interacts with the transcriptional regulator, CadC, to activate expression of the cadAB operon. This operon encodes the lysine decarboxylase, CadA, which converts lysine to cadaverine while consuming a cytoplasmic proton, and the antiporter, CadB, which exports protonated cadaverine in exchange for extracellular lysine. Together, these processes contribute to cytoplasmic pH homeostasis and support bacterial acid resistance - a mechanism essential for the survival of pathogenic bacteria in acidic host environments. Here, we present the cryo-EM structure of LysP from Pseudomonas aeruginosa in an inward-occluded conformation (3.2-5.3 Å resolution), bound to L-lysine and a nanobody. L-Lysine is coordinated by hydrophobic contacts, cation-π interactions, and by hydrogen bonding mostly with polar uncharged residues. Reconstitution of LysP into proteoliposomes confirms specific L-lysine transport, which is competitively inhibited by L-4-thialysine. These findings provide a structural framework for understanding selective lysine recognition and inhibition, with implications for antibacterial drug design.
- BioStruct-Africa, Nairobi, Kenya.
Organizational Affiliation: