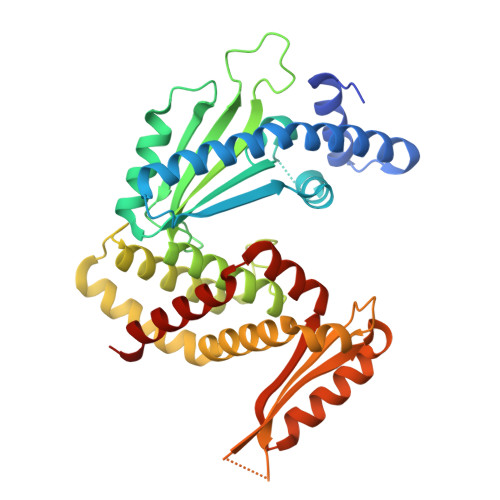
A bacterial immunity protein directly senses two disparate phage proteins.
Zhang, T., Cepauskas, A., Nadieina, A., Thureau, A., Coppieters 't Wallant, K., Martens, C., Lim, D.C., Garcia-Pino, A., Laub, M.T.(2024) Nature 635: 728-735
- PubMed: 39415022
- DOI: https://doi.org/10.1038/s41586-024-08039-y
- Primary Citation of Related Structures:
9AXB, 9ERV - PubMed Abstract:
Eukaryotic innate immune systems use pattern recognition receptors to sense infection by detecting pathogen-associated molecular patterns, which then triggers an immune response. Bacteria have similarly evolved immunity proteins that sense certain components of their viral predators, known as bacteriophages 1-6 . Although different immunity proteins can recognize different phage-encoded triggers, individual bacterial immunity proteins have been found to sense only a single trigger during infection, suggesting a one-to-one relationship between bacterial pattern recognition receptors and their ligands 7-11 . Here we demonstrate that the antiphage defence protein CapRel SJ46 in Escherichia coli can directly bind and sense two completely unrelated and structurally different proteins using the same sensory domain, with overlapping but distinct interfaces. Our results highlight the notable versatility of an immune sensory domain, which may be a common property of antiphage defence systems that enables them to keep pace with their rapidly evolving viral predators. We found that Bas11 phages harbour both trigger proteins that are sensed by CapRel SJ46 during infection, and we demonstrate that such phages can fully evade CapRel SJ46 defence only when both triggers are mutated. Our work shows how a bacterial immune system that senses more than one trigger can help prevent phages from easily escaping detection, and it may allow the detection of a broader range of phages. More generally, our findings illustrate unexpected multifactorial sensing by bacterial defence systems and complex coevolutionary relationships between them and their phage-encoded triggers.
- Department of Biology, Massachusetts Institute of Technology, Cambridge, MA, USA.
Organizational Affiliation: