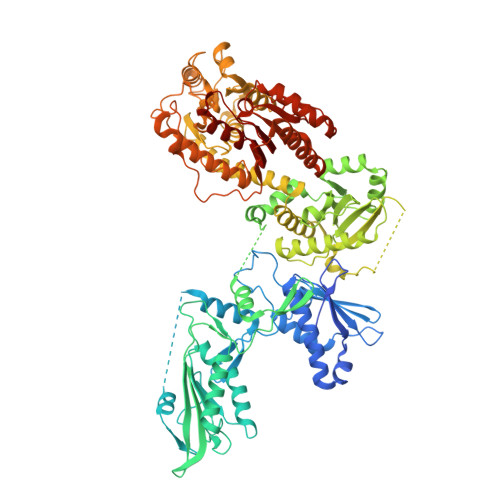
Phosphorylation-mediated conformational change regulates human SLFN11.
Kugler, M., Metzner, F.J., Witte, G., Hopfner, K.P., Lammens, K.(2024) Nat Commun 15: 10500-10500
- PubMed: 39627193
- DOI: https://doi.org/10.1038/s41467-024-54833-7
- Primary Citation of Related Structures:
9ERD, 9ERE, 9ERF, 9GMW, 9GMX - PubMed Abstract:
Human Schlafen 11 (SLFN11) is sensitizing cells to DNA damaging agents by irreversibly blocking stalled replication forks, making it a potential predictive biomarker in chemotherapy. Furthermore, SLFN11 acts as a pattern recognition receptor for single-stranded DNA (ssDNA) and functions as an antiviral restriction factor, targeting translation in a codon-usage-dependent manner through its endoribonuclease activity. However, the regulation of the various SLFN11 functions and enzymatic activities remains enigmatic. Here, we present cryo-electron microscopy (cryo-EM) structures of SLFN11 bound to tRNA-Leu and tRNA-Met that give insights into tRNA binding and cleavage, as well as its regulation by phosphorylation at S219 and T230. SLFN11 phosphomimetic mutant S753D adopts a monomeric conformation, shows ATP binding, but loses its ability to bind ssDNA and shows reduced ribonuclease activity. Thus, the phosphorylation site S753 serves as a conformational switch, regulating SLFN11 dimerization, as well as ATP and ssDNA binding, while S219 and T230 regulate tRNA recognition and nuclease activity.
- Gene Center and Department of Biochemistry, Ludwig-Maximilians-Universität München, Feodor-Lynen Straße 25, 81377, Munich, Germany.
Organizational Affiliation: