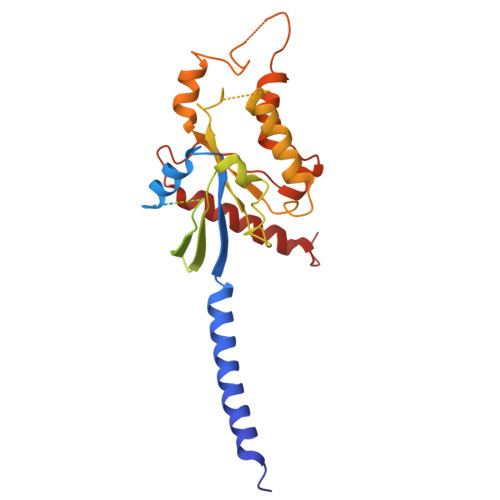
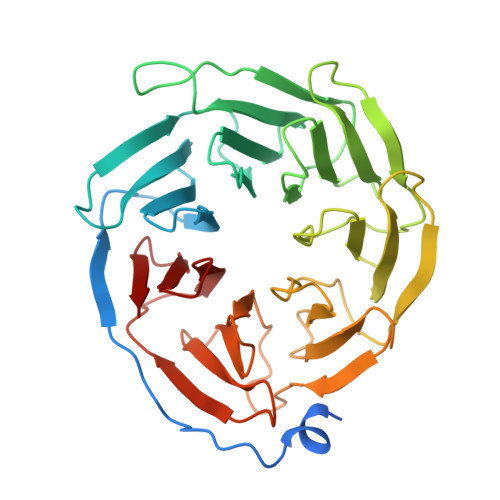
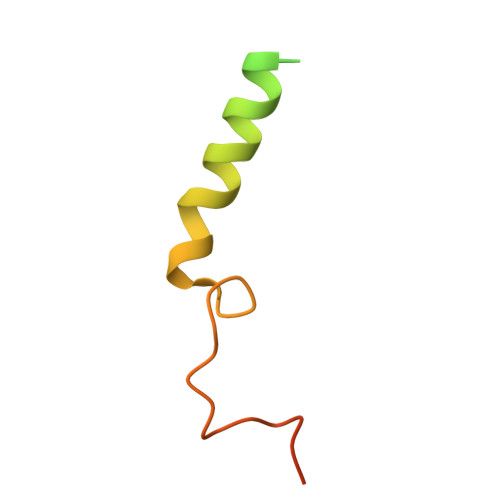
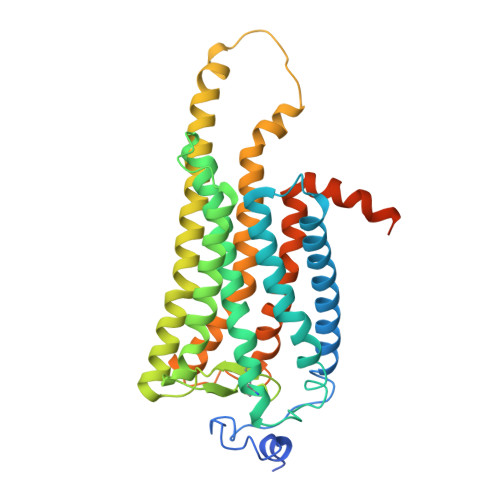
Active state structures of a bistable visual opsin bound to G proteins.
Tejero, O., Pamula, F., Koyanagi, M., Nagata, T., Afanasyev, P., Das, I., Deupi, X., Sheves, M., Terakita, A., Schertler, G.F.X., Rodrigues, M.J., Tsai, C.J.(2024) Nat Commun 15: 8928-8928
- PubMed: 39414813
- DOI: https://doi.org/10.1038/s41467-024-53208-2
- Primary Citation of Related Structures:
9EPP, 9EPQ, 9EPR - PubMed Abstract:
Opsins are G protein-coupled receptors (GPCRs) that have evolved to detect light stimuli and initiate intracellular signaling cascades. Their role as signal transducers is critical to light perception across the animal kingdom. Opsins covalently bind to the chromophore 11-cis retinal, which isomerizes to the all-trans isomer upon photon absorption, causing conformational changes that result in receptor activation. Monostable opsins, responsible for vision in vertebrates, release the chromophore after activation and must bind another retinal molecule to remain functional. In contrast, bistable opsins, responsible for non-visual light perception in vertebrates and for vision in invertebrates, absorb a second photon in the active state to return the chromophore and protein to the inactive state. Structures of bistable opsins in the activated state have proven elusive, limiting our understanding of how they function as bidirectional photoswitches. Here we present active state structures of a bistable opsin, jumping spider rhodopsin isoform-1 (JSR1), in complex with its downstream signaling partners, the G i and G q heterotrimers. These structures elucidate key differences in the activation mechanisms between monostable and bistable opsins, offering essential insights for the rational engineering of bistable opsins into diverse optogenetic tools to control G protein signaling pathways.
- Laboratory of Biomolecular Research, PSI Center for Life Sciences, Villigen-PSI, Switzerland.
Organizational Affiliation: