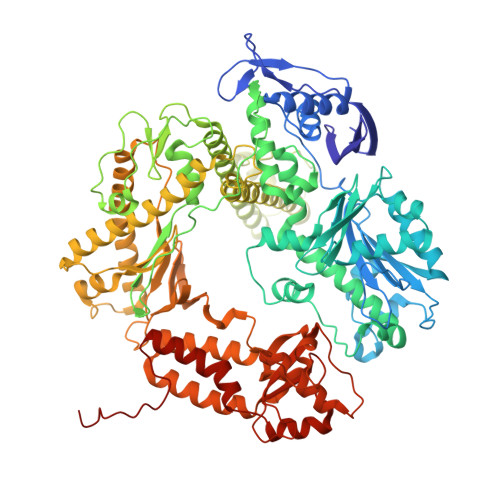
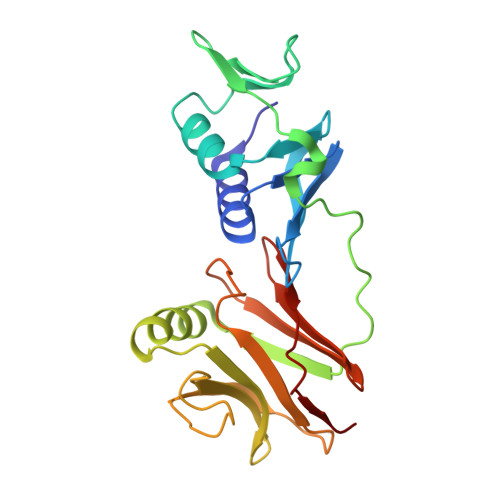
Structural insights into the exchange mechanism of a replicative DNA polymerase.
Feng, X., Spiering, M.M., Ruda de Luna Santos, A., Benkovic, S.J., Li, H.(2025) Nucleic Acids Res 53
- PubMed: 41459747
- DOI: https://doi.org/10.1093/nar/gkaf1359
- Primary Citation of Related Structures:
9E5Y, 9EA2, 9EA3, 9EA6 - PubMed Abstract:
Replicative DNA polymerases are distinguished by their speed, processivity, and fidelity. While speed and fidelity arise from the polymerase's intrinsic catalytic and proofreading activities, processivity is typically attributed to the DNA sliding clamp that tethers the polymerase to DNA. However, additional mechanisms may also contribute. The T4 bacteriophage polymerase can exchange on-the-fly, a process likely contributing to its ∼10-fold higher synthesis rate compared with human polymerases. Here, we reconstituted the T4 holoenzyme and polymerase exchange complexes using purified gp43 polymerase, gp45 sliding clamp, and a primer-template DNA substrate. Cryo-electron microscopy (cryo-EM) analysis revealed either one or two polymerases bound to the clamp and DNA. In the one-polymerase complex, the DNA threads perpendicularly through the clamp, supporting processive synthesis. In contrast, the two-polymerase complex displays a markedly tilted DNA orientation, impeding sliding and representing exchange intermediates. Three distinct conformational states of the two-polymerase complex define a multistep exchange mechanism. To our knowledge, these findings provide the first molecular-level view of replicative polymerase exchange.
- Department of Structural Biology, Van Andel Institute, Grand Rapids, MI 49503, United States.
Organizational Affiliation: