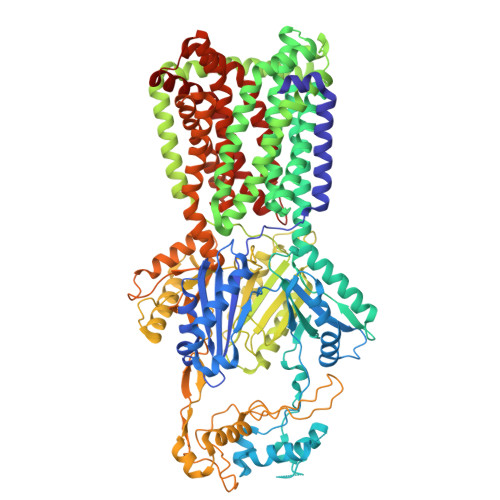
Cryo-EM structure of the Pseudomonas aeruginosa MexY multidrug efflux pump.
Gregor, W.D., Maharjan, R., Zhang, Z., Chiaraviglio, L., Sastry, N., Cui, M., Kirby, J.E., Yu, E.W.(2025) mBio 16: e0382624-e0382624
- PubMed: 40042268
- DOI: https://doi.org/10.1128/mbio.03826-24
- Primary Citation of Related Structures:
9E9F - PubMed Abstract:
Pseudomonas aeruginosa , a Gram-negative pathogen, has emerged as one of the most highly antibiotic-resistant bacteria worldwide and subsequently has become a leading cause of healthcare-associated, life-threatening infections. P. aeruginosa multidrug efflux Y (MexY) is an efflux pump that belongs to the resistance-nodulation-cell division (RND) superfamily. It is a major determinant for resistance to aminoglycosides in this opportunistic pathogen. However, the detailed molecular mechanisms involved in aminoglycoside recognition and extrusion by MexY have not been elucidated. Here, we report the cryo-electron microscopy structure of MexY to a resolution of 3.63 Å. The structure directly indicates two plausible pathways for drug export. It also suggests that MexY is capable of picking up antibiotics via the ceiling of the central cavity formed by the MexY trimer. Molecular dynamics simulations depict that MexY is able to use a tunnel connecting the central cavity to the funnel of the trimer to export its substrates. Here, we report the cryo-electron microscopy structure of the MexY multidrug efflux pump, posing the possibility that this pump is capable of capturing antibiotics from both the central cavity and the periplasmic cleft of the pump. The results indicate that MexY may utilize charged residues to bind and export drugs, mediating resistance to these antibiotics.
- Department of Pharmacology, Case Western Reserve University School of Medicine, Cleveland, Ohio, USA.
Organizational Affiliation: