Human NPR-A bound to ANP and XX16
Liu, S., Huang, X.-Y.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Atrial natriuretic peptide receptor 1 | 1,029 | Homo sapiens | Mutation(s): 0 Gene Names: NPR1, ANPRA EC: 4.6.1.2 | 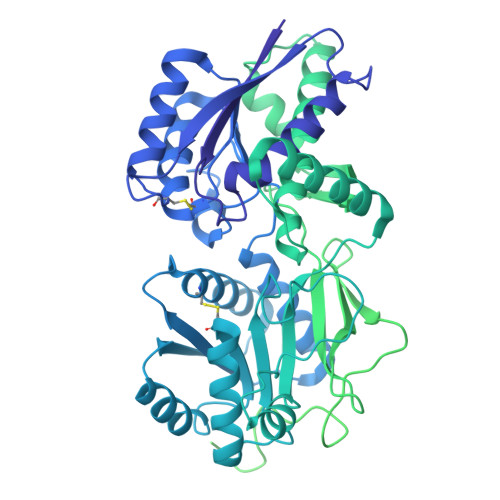 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P16066 (Homo sapiens) Explore P16066 Go to UniProtKB: P16066 | |||||
PHAROS: P16066 GTEx: ENSG00000169418 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P16066 | ||||
Glycosylation | |||||
| Glycosylation Sites: 2 | Go to GlyGen: P16066-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Atrial natriuretic peptide | 28 | Homo sapiens | Mutation(s): 0 Gene Names: NPPA, ANP, PND | 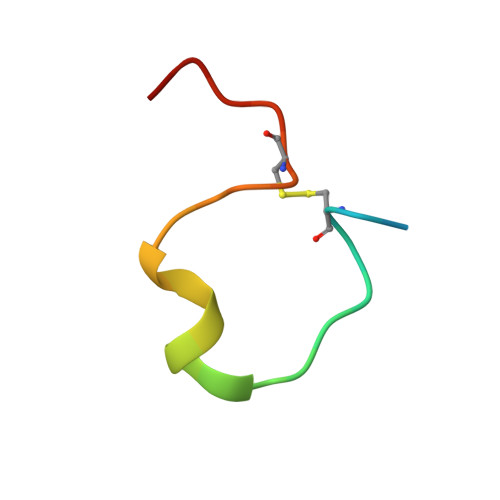 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P01160 (Homo sapiens) Explore P01160 Go to UniProtKB: P01160 | |||||
PHAROS: P01160 GTEx: ENSG00000175206 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P01160 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| XX16 - Heavy chain | D [auth H] | 228 | Mus musculus | Mutation(s): 0 | 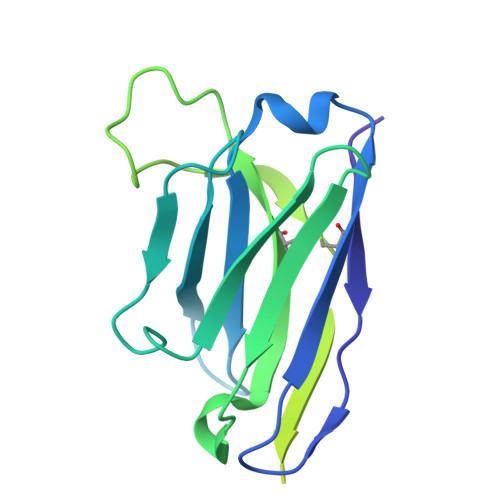 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| XX16 - Light chain | E [auth L] | 214 | Mus musculus | Mutation(s): 0 | 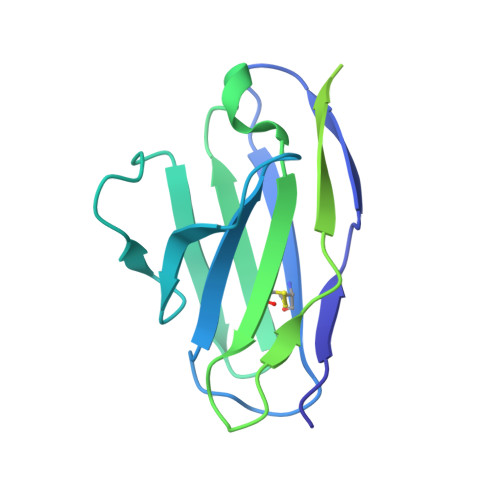 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CL (Subject of Investigation/LOI) Query on CL | J [auth A], K [auth B] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.20.1_4487: |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | -- |