Computational design of bifaceted protein nanomaterials with tailorable properties.
Rankovic, S., Carr, K.D., Decarreau, J., Skotheim, R., Kibler, R.D., Ols, S., Lee, S., Chun, J., Tooley, M., Dauparas, J., Eisenach, H.E., Glogl, M., Weidle, C., Borst, A.J., Baker, D., King, N.P.(2024) bioRxiv
- PubMed: 39484564
- DOI: https://doi.org/10.1101/2024.10.18.619149
- Primary Citation of Related Structures:
9DZE - PubMed Abstract:
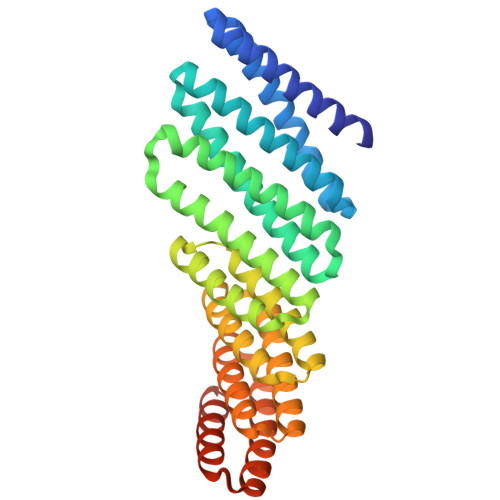
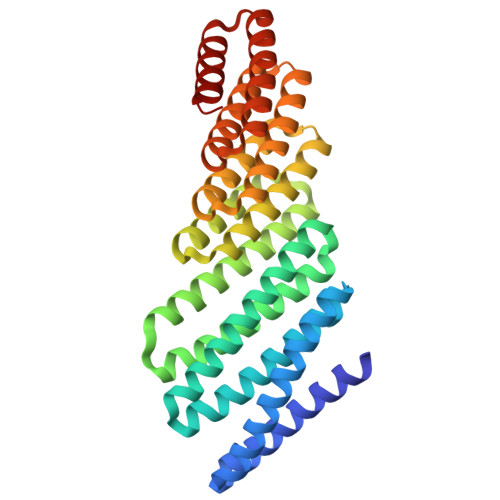
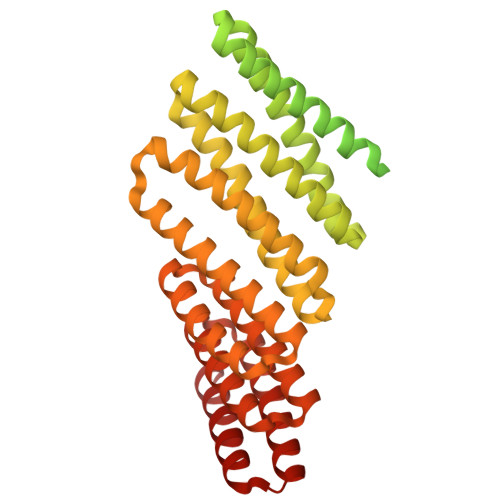
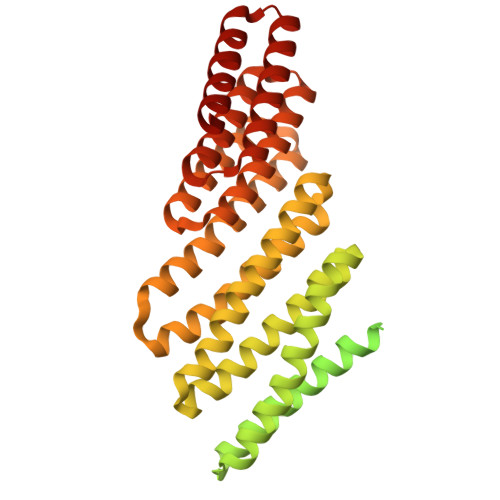
Recent advances in computational methods have led to considerable progress in the design of self-assembling protein nanoparticles. However, nearly all nanoparticles designed to date exhibit strict point group symmetry, with each subunit occupying an identical, symmetrically related environment. This property limits the structural diversity that can be achieved and precludes anisotropic functionalization. Here, we describe a general computational strategy for designing multi-component bifaceted protein nanomaterials with two distinctly addressable sides. The method centers on docking pseudosymmetric heterooligomeric building blocks in architectures with dihedral symmetry and designing an asymmetric protein-protein interface between them. We used this approach to obtain an initial 30-subunit assembly with pseudo-D5 symmetry, and then generated an additional 15 variants in which we controllably altered the size and morphology of the bifaceted nanoparticles by designing de novo extensions to one of the subunits. Functionalization of the two distinct faces of the nanoparticles with de novo protein minibinders enabled specific colocalization of two populations of polystyrene microparticles coated with target protein receptors. The ability to accurately design anisotropic protein nanomaterials with precisely tunable structures and functions will be broadly useful in applications that require colocalizing two or more distinct target moieties.
- Department of Biochemistry, University of Washington, Seattle, WA, USA.
Organizational Affiliation: