Structural Basis of Directionality Control in Large Serine Integrases
Shin, H.S., Rice, P.A.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Resolvase homolog YokA,SPbeta prophage-derived uncharacterized protein YotN | 620 | Bacillus subtilis | Mutation(s): 0 Gene Names: yokA, BSU21660, yotN, yokJ, BSU19820 | 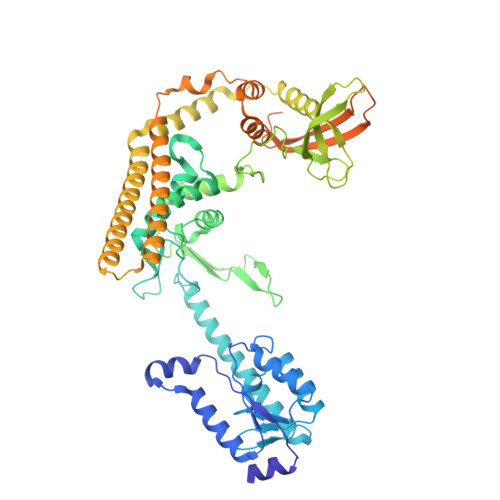 | |
UniProt | |||||
Find proteins for O32006 (Bacillus subtilis (strain 168)) Explore O32006 Go to UniProtKB: O32006 | |||||
Find proteins for O34850 (Bacillus subtilis (strain 168)) Explore O34850 Go to UniProtKB: O34850 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Groups | O34850O32006 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (34-MER) | 34 | Bacillus subtilis |  | ||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (33-MER) | 33 | Bacillus subtilis |  | ||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (25-MER) | 25 | Bacillus subtilis |  | ||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (24-MER) | 24 | Bacillus subtilis |  | ||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ZN (Subject of Investigation/LOI) Query on ZN | Q [auth A], R [auth C], S [auth I], T [auth K] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.21.1_5286: |
| RECONSTRUCTION | cryoSPARC |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Science Foundation (NSF, United States) | United States | -- |