Molecular Basis for Peptide Nitration by a Novel Cytochrome P450 Enzyme in RiPP Biosynthesis.
Nolan, K., Usai, R., Li, B., Jordan, S., Wang, Y.(2025) ACS Catal 15: 10391-10404
- PubMed: 40568218
- DOI: https://doi.org/10.1021/acscatal.5c01932
- Primary Citation of Related Structures:
9DUJ, 9EBY - PubMed Abstract:
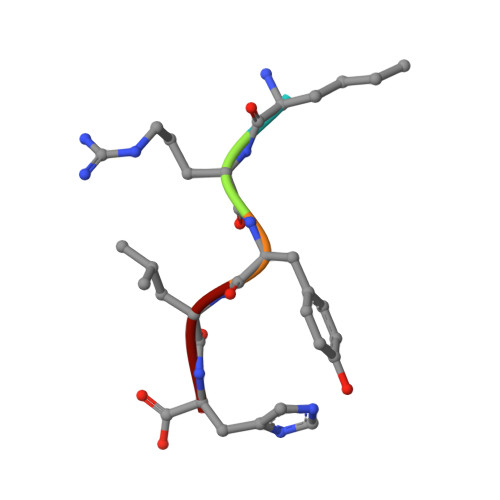
RufO is a unique cytochrome P450 enzyme (CYP) involved in the biosynthesis of rufomycin, an antituberculosis cyclic peptide featuring an unusual nitrated tyrosine. Recent studies have clarified RufO's role in producing ribosomally synthesized and post-translationally modified peptides (RiPPs). Despite growing interest in nitrating enzymes and RiPP biosynthesis, the mechanism by which RufO recognizes and nitrates its pentapeptide substrate, MRYLH, remains poorly understood. In this study, we use a combination of spectroscopic, kinetic, and structural techniques to elucidate the molecular basis for peptide binding and heme-based nitration in RufO. Peptide binding is an endothermic process with a dissociation constant of 0.78 μM. Unlike most CYPs, RufO does not undergo the typical spin state conversion nor exhibit a significant increase in reduction potential upon substrate binding. The minimal perturbation to the heme center may lead to RufO's lack of specificity for redox partners. However, significant shifts in the vibrational frequencies of carbonyl complexes upon substrate binding indicate a more polar heme distal site that favors a nonlinear binding conformation of diatomic gas molecules. These distinctive features contrast with TxtE, the only other CYP known to catalyze aromatic nitration. A 1.51 Å resolution crystal structure reveals that substrate binding induces significant conformational changes in the distal pocket, particularly in the regions interacting with Arg-2 and His-5 of the MRYLH peptide. While Tyr-3 is positioned similarly to its counterpart in P450 Blt , a paralog that catalyzes peptide cross-linking, an extended hydrogen-bonding network constraining His-5 is unique to RufO and likely contributes to its distinct nitration activity. Furthermore, transient kinetic data suggest the sequential binding of O 2 followed by • NO and characterize a ferric-superoxo intermediate essential for the nitration activity. This study provides valuable insights into the substrate specificity and catalytic mechanisms of CYPs involved in nitration reactions and RiPP biosynthesis.
- Department of Chemistry, University of Georgia, Athens, Georgia 30602, United States.
Organizational Affiliation: