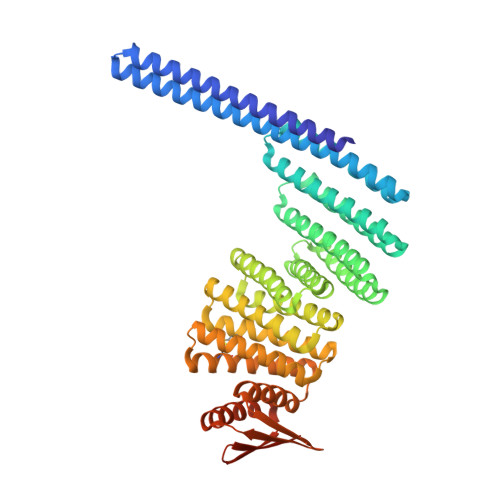
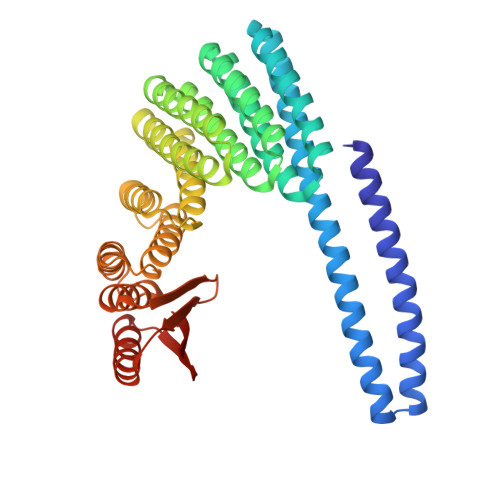
Bond-centric modular design of protein assemblies.
Wang, S., Favor, A., Kibler, R.D., Lubner, J.M., Borst, A.J., Coudray, N., Redler, R.L., Chiang, H.T., Sheffler, W., Hsia, Y., Bethel, N.P., Li, Z., Ekiert, D.C., Bhabha, G., Pozzo, L.D., Baker, D.(2025) Nat Mater 24: 1644-1652
- PubMed: 40745093
- DOI: https://doi.org/10.1038/s41563-025-02297-5
- Primary Citation of Related Structures:
9DRL - PubMed Abstract:
Directional interactions that generate regular coordination geometries are a powerful means of guiding molecular and colloidal self-assembly, but implementing such high-level interactions with proteins remains challenging due to their complex shapes and intricate interface properties. Here we describe a modular approach to protein nanomaterial design inspired by the rich chemical diversity that can be generated from the small number of atomic valencies. We design protein building blocks using deep learning-based generative tools, incorporating regular coordination geometries and tailorable bonding interactions that enable the assembly of diverse closed and open architectures guided by simple geometric principles. Experimental characterization confirms the successful formation of more than 20 multicomponent polyhedral protein cages, two-dimensional arrays and three-dimensional protein lattices, with a high (10%-50%) success rate and electron microscopy data closely matching the corresponding design models. Due to modularity, individual building blocks can assemble with different partners to generate distinct regular assemblies, resulting in an economy of parts and enabling the construction of reconfigurable networks for designer nanomaterials.
- Department of Biochemistry, University of Washington, Seattle, WA, USA. swang523@uw.edu.
Organizational Affiliation: