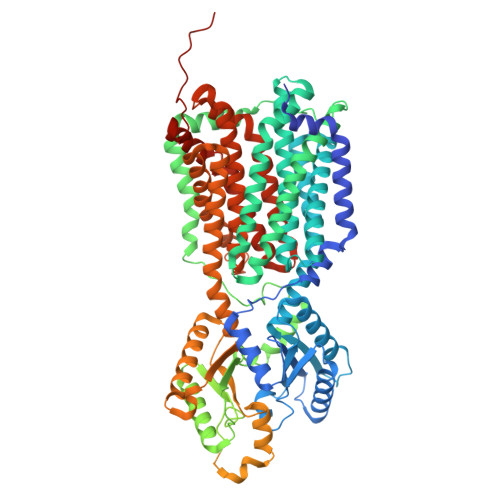
Structures of the mycobacterial MmpL4 and MmpL5 transporters provide insights into their role in siderophore export and iron acquisition.
Maharjan, R., Zhang, Z., Klenotic, P.A., Gregor, W.D., Tringides, M.L., Cui, M., Purdy, G.E., Yu, E.W.(2024) PLoS Biol 22: e3002874-e3002874
- PubMed: 39423221
- DOI: https://doi.org/10.1371/journal.pbio.3002874
- Primary Citation of Related Structures:
9B43, 9B46, 9DP6 - PubMed Abstract:
The Mycobacterium tuberculosis (Mtb) pathogen, the causative agent of the airborne infection tuberculosis (TB), harbors a number of mycobacterial membrane protein large (MmpL) transporters. These membrane proteins can be separated into 2 distinct subclasses, where they perform important functional roles, and thus, are considered potential drug targets to combat TB. Previously, we reported both X-ray and cryo-EM structures of the MmpL3 transporter, providing high-resolution structural information for this subclass of the MmpL proteins. Currently, there is no structural information available for the subclass associated with MmpL4 and MmpL5, transporters that play a critical role in iron homeostasis of the bacterium. Here, we report cryo-EM structures of the M. smegmatis MmpL4 and MmpL5 transporters to resolutions of 2.95 Å and 3.00 Å, respectively. These structures allow us to propose a plausible pathway for siderophore translocation via these 2 transporters, an essential step for iron acquisition that enables the survival and replication of the mycobacterium.
- Department of Pharmacology, Case Western Reserve University School of Medicine, Cleveland, Ohio, United States of America.
Organizational Affiliation: