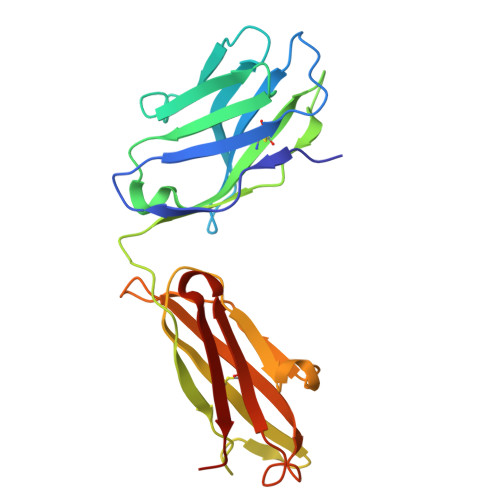
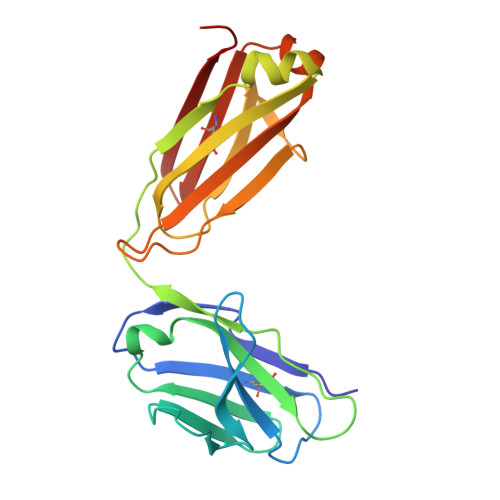
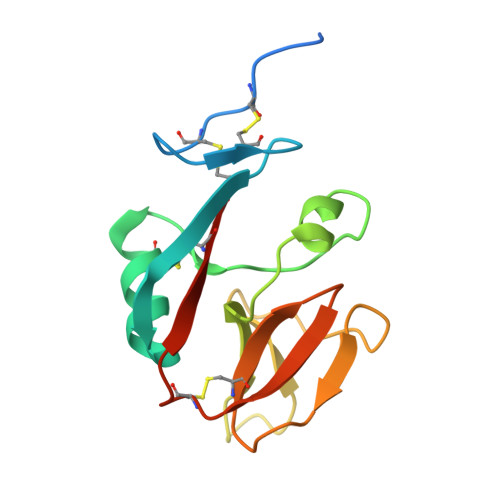
Agonistic anti-NKG2D antibody structure reveals unique stoichiometry and epitope compared to natural ligands.
Fallon, D., Huang, C.S., Ma, J., Morgan, C., Zhou, Z.S.(2024) MAbs 16: 2433121-2433121
- PubMed: 39582357
- DOI: https://doi.org/10.1080/19420862.2024.2433121
- Primary Citation of Related Structures:
9DH2 - PubMed Abstract:
Natural killer (NK) cells are effector cells of the innate immune system that distinguish between healthy and abnormal cells through activating and inhibitory receptor signaling. NKG2D, a homodimeric activating receptor expressed on NK cells, recognizes a diverse class of stress ligands expressed by cells experiencing infection, malignant transformation, chronic inflammation, and other cellular stresses. Despite the variety of NKG2D ligands, they all bind the receptor asymmetrically in a 1:1 ligand to homodimeric NKG2D stoichiometry. In contrast, as we report herein, the agonistic antibody 2D3 binds NKG2D with a 2:1 stoichiometry of its antigen binding fragments to homodimeric NKG2D and a largely distinct epitope. This binding interaction, as compared to NKG2D natural ligands, suggests there may be unique mechanisms to engage this receptor while offering possible benefits when incorporated into an IgG-based therapeutic.
- Dragonfly Therapeutics, Inc., Waltham, MA, USA.
Organizational Affiliation: