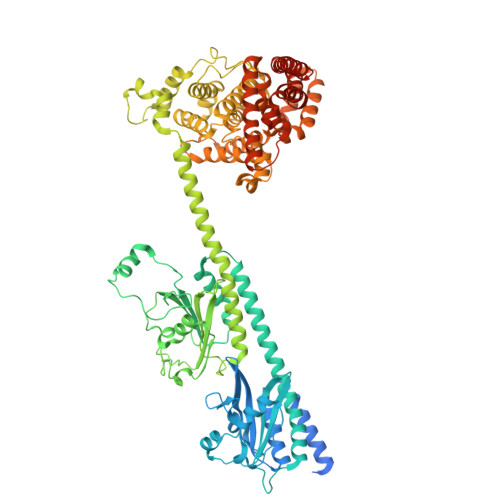
Structural and functional dynamics of human cone cGMP-phosphodiesterase important for photopic vision.
Singh, S., Srivastava, D., Boyd, K., Artemyev, N.O.(2025) Proc Natl Acad Sci U S A 122: e2419732121-e2419732121
- PubMed: 39739818
- DOI: https://doi.org/10.1073/pnas.2419732121
- Primary Citation of Related Structures:
9CXG, 9CXH, 9CXI, 9CXJ - PubMed Abstract:
Cone cGMP-phosphodiesterase (PDE6) is the key effector enzyme for daylight vision, and its properties are critical for shaping distinct physiology of cone photoreceptors. We determined the structures of human cone PDE6C in various liganded states by single-particle cryo-EM that reveal essential functional dynamics and adaptations of the enzyme. Our analysis exposed the dynamic nature of PDE6C association with its regulatory γ-subunit (Pγ) which allows openings of the catalytic pocket in the absence of phototransduction signaling, thereby controlling photoreceptor noise and sensitivity. We demonstrate evolutionarily recent adaptations of PDE6C stemming from residue substitutions in the Pγ subunit and the noncatalytic cGMP binding site and influencing the Pγ dynamics in holoPDE6C. Thus, our structural analysis sheds light on the previously unrecognized molecular evolution of the effector enzyme in cones that advances adaptation for photopic vision.
- Department of Molecular Physiology and Biophysics, University of Iowa Carver College of Medicine, Iowa City, IA 52242.
Organizational Affiliation: