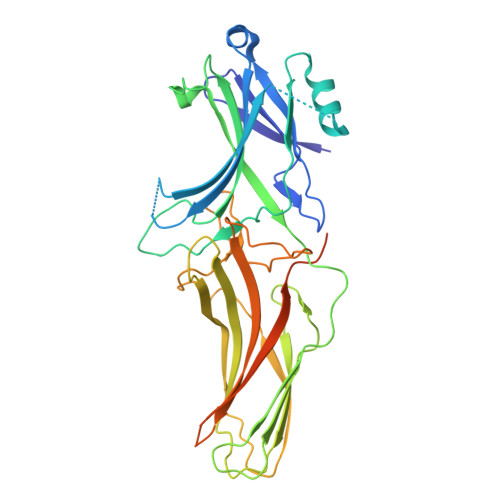
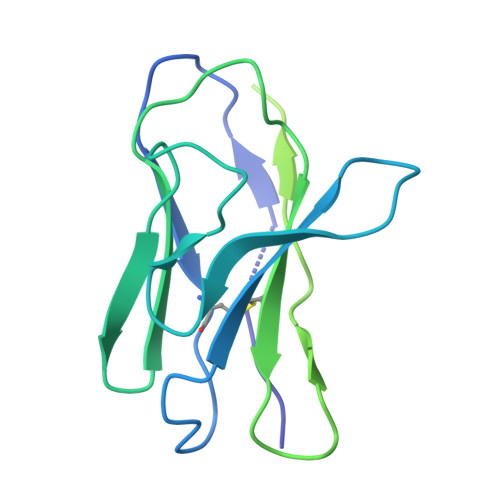
Mechanism of beta-arrestin 1 mediated Src activation via Src SH3 domain revealed by cryo-electron microscopy.
Pakharukova, N., Thomas, B.N., Bansia, H., Li, L., Bassford, D.K., Abzalimov, R.R., Kim, J., Kahsai, A.W., Pani, B., Xiao, K., Ochakovski, R., Liu, S., Zhang, X., Ahn, S., des Georges, A., Lefkowitz, R.J.(2025) bioRxiv
- PubMed: 39131402
- DOI: https://doi.org/10.1101/2024.07.31.605623
- Primary Citation of Related Structures:
9BT8, 9CX3, 9CX9 - PubMed Abstract:
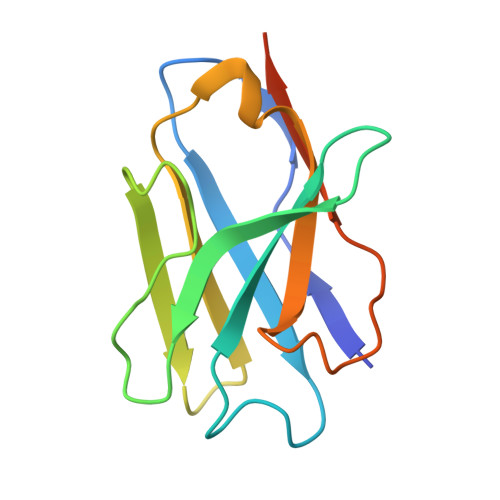
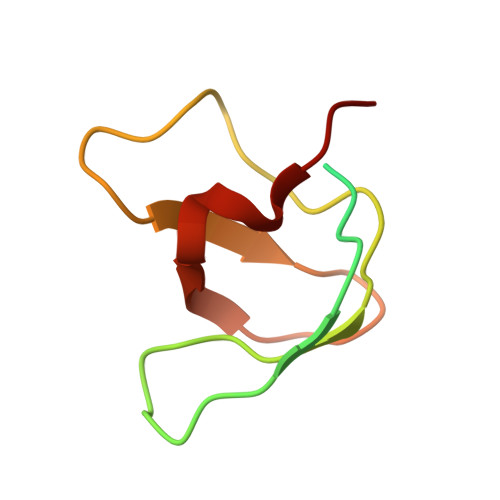
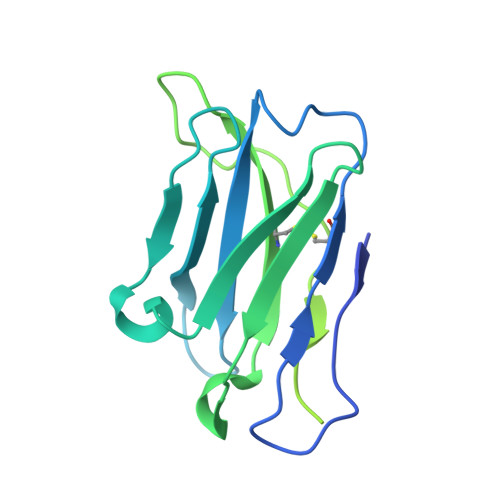
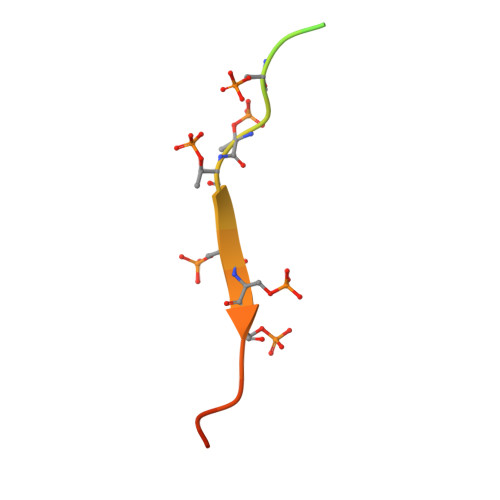
Beta-arrestins (βarrs) are key regulators and transducers of G-protein coupled receptor signaling; however, little is known of how βarrs communicate with their downstream effectors. Here, we use cryo-electron microscopy to elucidate how βarr1 recruits and activates non-receptor tyrosine kinase Src. βarr1 binds Src SH3 domain via two distinct sites: a polyproline site in the N-domain and a non-proline site in the central crest region. At both sites βarr1 interacts with the aromatic surface of SH3 which is critical for Src autoinhibition, suggesting that βarr1 activates Src by SH3 domain displacement. Binding of SH3 to the central crest region induces structural rearrangements in the β-strand V, finger, and middle loops of βarr1 and interferes with βarr1 coupling to the receptor core potentially impacting receptor desensitization and downstream signaling.
- Department of Medicine, Duke University Medical Center; Durham, NC 27710, USA.
Organizational Affiliation: