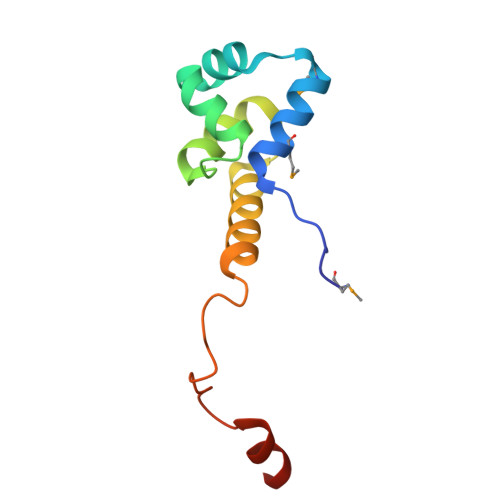
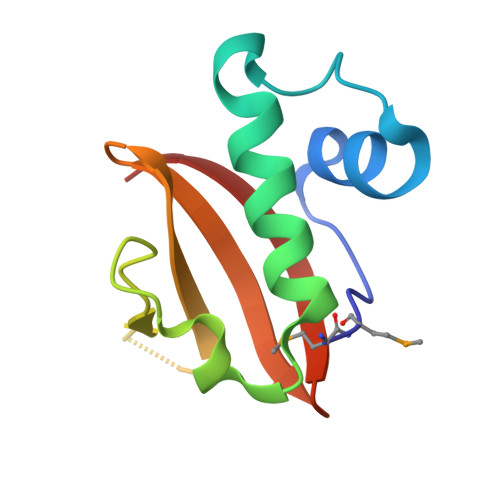
Antitoxin control of optimal transcriptional repression in the atypical HigB-HigA toxin-antitoxin system from Proteus vulgaris.
Pavelich, I.J., Schureck, M.A., Srinivas, P., Blackburn, T.M., Wang, D., Hoffer, E.D., Boamah, M., Zaldana, K., Onuoha, N., Miles, S.J., Grabowicz, M., Okafor, C.D., Dunham, C.M.(2025) Nucleic Acids Res 53
- PubMed: 40671524
- DOI: https://doi.org/10.1093/nar/gkaf610
- Primary Citation of Related Structures:
9CHL, 9CHN - PubMed Abstract:
Bacterial toxin-antitoxin (TA) pairs transcriptionally autoregulate their expression via a repression/derepression mechanism in response to changing environmental conditions. The structural diversity of TA systems influences the mechanisms of transcriptional regulation. Here, we define the molecular mechanism for the plasmid-encoded HigB-HigA TA pair originally identified in a post-operative infection with antibiotic-resistant Proteus vulgaris. We determine DNA binding and promoter activity by the HigB-HigA complex supported by structural biology and molecular dynamics simulations of an elusive DNA operator-TA repressor complex. To define the optimal oligomeric TA repressor-DNA operator complex required for derepression, we engineered a dedicated trimeric HigB-HigA2 complex that represses transcription more than 26-fold as compared to the tetrameric HigB2-HigA2. These results expand the known diversity of how the HigB-HigA TA family is autoregulated.
- Department of Chemistry and the Emory Antibiotic Resistance Center, Emory University, Atlanta, GA 30322, United States.
Organizational Affiliation: