The Structure of Spiroplasma Virus 4 : Exploring the Capsid Diversity of the Microviridae.
Mietzsch, M., Kailasan, S., Bennett, A., Chipman, P., Fane, B., Huiskonen, J.T., Clarke, I.N., McKenna, R.(2024) Viruses 16
- PubMed: 39066266
- DOI: https://doi.org/10.3390/v16071103
- Primary Citation of Related Structures:
9CGM - PubMed Abstract:
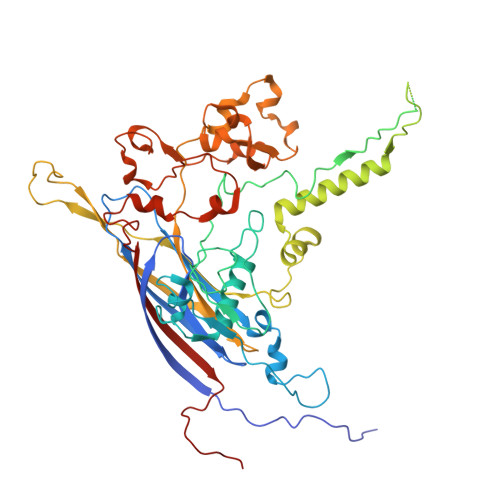
Spiroplasma virus 4 (SpV4) is a bacteriophage of the Microviridae , which packages circular ssDNA within non-enveloped T = 1 icosahedral capsids. It infects spiroplasmas, which are known pathogens of honeybees. Here, the structure of the SpV4 virion is determined using cryo-electron microscopy to a resolution of 2.5 Å. A striking feature of the SpV4 capsid is the mushroom-like protrusions at the 3-fold axes, which is common among all members of the subfamily Gokushovirinae. While the function of the protrusion is currently unknown, this feature varies widely in this subfamily and is therefore possibly an adaptation for host recognition. Furthermore, on the interior of the SpV4 capsid, the location of DNA-binding protein VP8 was identified and shown to have low structural conservation to the capsids of other viruses in the family. The structural characterization of SpV4 will aid future studies analyzing the virus-host interaction, to understand disease mechanisms at a molecular level. Furthermore, the structural comparisons in this study, including a low-resolution structure of the chlamydia phage 2, provide an overview of the structural repertoire of the viruses in this family that infect various bacterial hosts, which in turn infect a wide range of animals and plants.
- Department of Biochemistry and Molecular Biology, College of Medicine, Center for Structural Biology, McKnight Brain Institute, University of Florida, Gainesville, FL 32610, USA.
Organizational Affiliation: